DNA N6-methyladenine (6mA) modification is common in prokaryotes1and eukaryotes, involving in gene regulation, transposon, stem cell differentiation, and human tumors. hALKBH1 exhibits demethylation activity toward 6mA, and its abnormal expression has been found in many human cancers and developmental defects, such as tissue malformation and gender imbalance. DNA methyltransferase N6AMT1 and demethylase hALKBH1 mediate the methylation and demethylation of DNA 6mA in the human genome, respectively. The abnormal distribution of 6mA has been found in many cancers. But, the molecular function of hALKBH1 is still controversial and the functional mechanism is unclear.
In a new study published on Mar. 10th in Cell Research, led by XU Wenqing / LIANG Dong-Cai’s group from the Institute of Biophysics (IBP), collaborated with CHEN Zhongzhou’s group from China Agricultural University, researchers solved the two high-resolution crystal structures of binary and ternary complex of human Alkbh1. Compared with other AlkB family proteins, Alkbh1 displays several novel structural features, especially a unique region Flip0 on top of the conserved double-stranded β-helix fold. The distinct Flip0 plays a crucial role in substrate recognition and catalytic m6A demethylase activity of Alkbh1. Differentiation between single and double strand DNA by Flip0 is a special substrate selection mechanism found in Alkbh1; and a very long and positive Flip1, there are a large number of basic residues on the side facing the active pocket, forming a large positive charge region; mutation of these residues will make ALKBH1 lose its ability to bind to DNA. Flip2 is similar to other members of the AlkB family in structure and contains a long loop. It is found that two residues Tyr177 and Trp179 on Flip2 participate in the recognition and binding of DNA-6mA through point mutation experiments. In addition, Tyr184 and Glu236 near the active center of ALKBH1 moved to the active center and formed a stable triangular structure with Arg344 through hydrogen bonds after binding to α-KG. The stable triangle formed near the binding site of α-KG is the support of ALKBH1 catalysis.
Researchers developed a novel, high-throughput and convenient enzymatic assay. Combined with structural analysis, reveal that unpairing dsDNA (Bulge/Bubble), rather than ssDNA or dsDNA, are preferential substrates of ALKBH1 and more important physiologically. As the second form of DNA modification, 6mA plays important roles in the epigenetic regulation of the mammalian genome. Consistent with these findings of the strong 6mA demethylation activity of ALKBH1 in Bulge/Bubble DNA, 6mA has also been found to exert functional roles in DNA mismatch repair. The studies provide a theoretical basis for the study of substrate selection mechanism, catalytic reaction mechanism and physiological function of ALKBH1, and the important basis for drug design of related diseases. 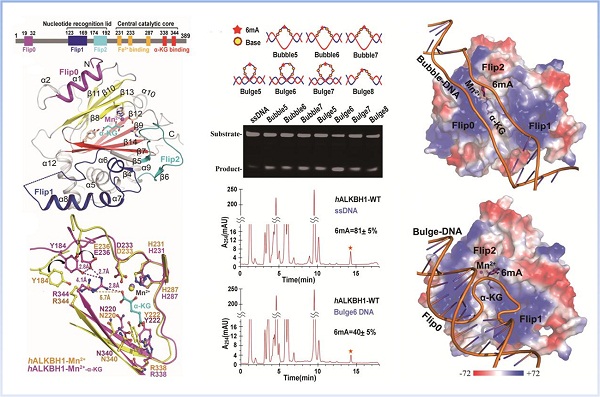
ALKBH1 prefers locally unpaired DNAs -Bubble/Bulge DNA 6mA as substrates
(The image by Dr. XU Wenqing/ LIANG Dong-Cai’s Lab)
The project was funded by the National Laboratory of Biomacromolecules at IBP, National Natural Science Foundation of China and the Strategic Priority Research Program of CAS. The staff of the BL17U1 and BL19U1 beamlines at the Shanghai Synchrotron Radiation Facility and the Science research platform at IBP have provided technical supports. TIAN Li-Fei, LIU Yan-Ping, CHEN Lianqi, TANG Qun and SUN Wei from XU Wenqing/ LIANG Dong-Cai’s group, WEI Wu from CHEN Zhongchou’s Group, etc participate in this work. Dr. YAN Xiao-Xue and Dr. CHEN Zhongchou are co-corresponding authors.
The web link for this paper is https://www.nature.com/articles/s41422-019-0233-9
Contact: YAN Xiao-Xue
Institute of Biophysics, Chinese Academy of Sciences
Beijing100101, China
Tel: (86)-10-64888509
Email: snow@ibp.ac.cn(Reported by Dr. XU Wenqing/ LIANG Dong-Cai’s group) |