The Calvin-Benson-Bassham (CBB) cycle (also termed as the dark reactions) is responsible for CO2 assimilation and carbohydrate production in oxygenic photosynthetic organisms. The CBB cycle is regulated by light/dark transitions through the redox states of chloroplast thioredoxins (TRXs). TRXs receive electrons from photosystem I upon illumination, further reduce and activate enzymes of CBB cycle. In addition, a small chloroplast protein CP12, which is central in the regulation of CBB cycle, is also redox-regulated by TRXs.
Phosphoribulokinase (PRK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are two essential enzymes of the CBB cycle, consuming the products of light reactions, namely ATP and NADPH, respectively. BothPRK and GAPDHare regulated by TRXs, being active at the reduced state and inactive when oxidized. Moreover, PRK and GAPDH are able to assemble with CP12 in the dark, forming GAPDH/CP12/PRK complex. The activities of both PRK and GAPDH are further inhibited within the complex.
On March. 8th, 2020, a research article entitled “Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle” was published in the PLANT CELL by Professor LI Mei’s group from the Institute of Biophysics, Chinese Academy of Sciences. They reported four crystal structures of PRK, including the structure of PRK from Synechococcus sp. PCC 7942 (SePRK) in complex with adenosine diphosphate (ADP) and glucose-6-phosphate (G6P), and the structure of GAPDH/CP12/PRK complex from Arabidopsis thaliana.
Based on the structural data, as well as the results of biochemical and enzymatic assays, theyidentifiedthe active site and residues involved in catalysisof PRK, unraveled the key elements essential for GAPDH/CP12/PRK complex assembly, and suggested the regulative mechanism of PRK and the CBB cycle.
This study provides structural basis for understanding thecatalytic and regulative mechanisms of PRK, andadvances our knowledge about the subtle regulations of the CBB cycle in response to the light/dark transitions.
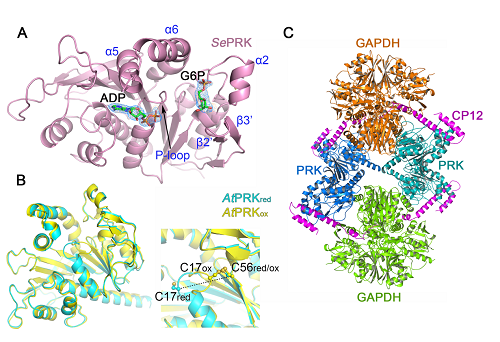
Figure Legend:
(A) Structure of SePRK complexed with ADP and G6P. Key elements for binding ligands are marked in blue. (B) Structure comparison of AtPRKred and AtPRKox. The P-loop conformation is different due to the C17-C56 disulfide bond formation in the AtPRKox. structure. (C) Structure of GAPDH/CP12/PRK complex. Two GAPDH tetramers (orange and lime-green), two PRK dimers (blue and teal) and four CP12 monomers (magenta) are shown in cartoon.
(The image by Dr. LI Mei’s lab)
The web link for this paper is http://www.plantcell.org/content/32/5/1556
Contact: LI Mei
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Phone: 86-10-64888511
Email: meili@ibp.ac.cn
(Reported by Dr. LI Mei's group)