On Apr. 6th, researchers from the groups of Professor ZHU Ping and Professor SUN Fei at the Institute of Biophysics, Chinese Academy of Sciences and their collaborators published online ahead of print a research article entitled “Cryo-EM structure of a transcribing cypovirus” in the Proceedings of the National Academy of Sciences (PNAS) (Yang et al, www.pnas.org/cgi/doi/10.1073/pnas.1200206109).
Taking advantage of the method of cryoEM single particle analysis, researchers reconstructed the 3D density map of a transcribing dsRNA virus, CPV, at the near-atomic resolution (4.1 Å), and built an atomic structural model of this transcribing virus. Following the paper entitled “Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping” (Chen et al, PNAS, 2011, 108:1373-1378) published by the same groups, this work resprsents a new progress made by the researchers in IBP in the field of 3D reconstruction of virus and biomacromolecules at the near-atomic resolution by cryoEM analysis and structural study of double-stranded RNA (dsRNA) viruses.
Since the viruses in the process of transcribing are metastable and difficult to be crystallized, no crystal structure of the transcribing virus has been reported. The cryoEM method provides a suitable tool to analyze the structure of viruses during transcribing process. However, the limited resolution (around 20 Å) of previously published cryoEM structural studies on transcribing dsRNA viruses make the mRNA transcription process and underlying molecular mechanism remain elusive.The newly published study made by Yant et al significantly improves the reconstruction resolution, to the near-atomic level for the first time, of the transcriping dsRNA virus structure. It represents the first reported reconstructed density map, with single amino acid residue readily visible, of dsRNA virus at the transcribing stage by cryoEM analysis.
This study revealed the structural changes of the capsid proteins that are associated with dsRNA virus transcription, which is an important stage in dsRNA virus life cycle. It not only provided structural evidence to the hypothesis of dsRNA virus mRNA release channel in virus capsid shell, but also observed the active sites of GTase in the capping state which was predicated previously by biochemical analyses. A highly coordinated, capsid proteins-involved, transcription and mRNA capping process of dsRNA virus was revealed. The results also provide insights to conclude the debates on the mRNA release channel of dsRNA virus.
This work was supported by grants from the Ministry of Science and Technology of China, the National Natural Science Foundation of China and the Chinese Academy of Sciences.
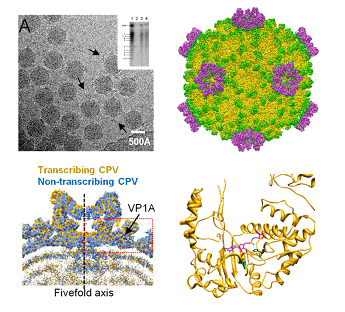
Figure legends: The upper-left panel shows a cryo-EM image of CPV recorded by CCD and the inserted PAGE image indicates that the viruses are transcribing. The upper-right panel shows the reconstructed density map of transcribing CPV. The left-bottom panel shows the differences of fivefold vertex between transcribing and nontranscrbing virus structure. The right-bottom panel shows the GTase domain of CPV and its linked substrase surrounded by related residues.