IBP Scientists Revealed The Molecular Mechanism for RNA-Guided RNA Cleavage by Cas13a
Almost all archaea and half of bacteria possess Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated genes (Cas) adaptive immune systems, which protect microbes from the foreign nucleic acids. Cas13a is a newly identified effector of Class 2 and type VI CRISPR-Cas system. Cas13a was found to be a single-component programmable RNA-guided RNA-targeting CRISPR effector, protecting the host from the RNA viruses. Upon recognition of its RNA target, activated Cas13a engages in “collateral” cleavage of any free RNA in solution. However, the molecular mechanism of RNA-guided RNA cleavage is unknown.
In the study published online in Cell on July 27, 2017, entitled The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a, LIU Liang, LI Xueyan, MA Jun, LI Zongqiang and colleagues (led by Prof. WANG Yanli and Prof. ZHANG Xinzheng at the institute of Biophysics of Chinese Academy of Sciences) solved the 3.08 ? crystal structure of Leptotrichia buccalis (Lbu) Cas13a bound to crRNA and its target RNA, as well as the 3.2 ? cryo-EM structure of the LbuCas13a-crRNA complex.
These structural studies revealed that LbuCas13a adopts a bilobed architecture consisting of an α-helical REC lobe and a NUC lobe. The crRNA and target RNA form duplex RNA, located in a positively-charged central channel of the NUC lobe. Interestingly, Cas13a protein and crRNA undergo significant conformational change upon target RNA binding. LIU Liang et al. found that the guild-target RNA duplex formation triggers the conserved catalytic residues in HEPN1 domain to move towards the HEPN2 domain, activating the HEPN catalytic site of Cas13a. The activated Cas13a cleaves any single-stranded RNA in solution in a sequence non-specific manner. Therefore, LIU's studies revealed a unique molecular mechanism by which Cas13a protein cleaves both single-stranded target RNA and collateral RNAs, resulting in cell death in bacteria. This altruistic cell suicide mediated by Cas13a is likely essential for evolution bacteria evolution.
Together with their earlier studies, LIU Liang et al. revealed that Cas13a has two separate catalytic sites for its two interdependent RNase activities. The HEPN1 domain and HEPN2 domain are responsible for RNA cleavage. The Helical-1 and HEPN2 domains may be also involved in pre-crRNA stabilization and processing. The HEPN2 domain appears to catalyze pre-crNA processing in Lbu, while the Helical-1 domain is critical for pre-crRNA processing in Leptotrichia shahii (Lsh).
These structural studies provide critical insights into the molecular mechanism of target RNA recognition and activation of Cas13a, which will significantly facilitate the research on this type VI CRISPR-Cas system and pave the way towards better development and utilizing of the RNA editing tool.
The research was funded by Natural Science Foundation of China (91440201, 31571335, 31400640, 31570874), the Chinese Ministry of Science and Technology (2014CB910102), and the Strategic Priority Research program of the Chinese Academy of Sciences (XDB08010203). The X-ray diffraction data were collected at beamlines BL-17U and BL-19U at the Shanghai Synchrotron Radiation Facility. The cryo-EM data were collected at the Center for Biological Imaging (IBP, CAS).
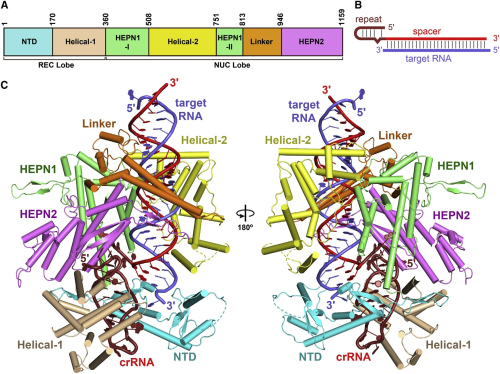
Figure 1. The Crystal Structure of LbuCas13a-crRNA-target RNA Ternary Complex (Image by IBP)
Contact:
WANG Yanli
Institute of Biophysics
Email:ylwang@ibp.ac.cn
Tel: 86-10-64881316
Fax:86-10-64881316

