SALL4A Regulates DNA Demethylation at Enhancers
DNA methylation is broadly recognized as the most stable epigenetic modification. Its distribution pattern in parental cells is faithfully transmitted to daughter cells during mitosis. Recent discoveries of the TET-mediated DNA oxidative demethylation greatly expanded our understanding about the plasticity and dynamics of this modification.
TET family enzymes (TET1, TET2 and TET3) could successively oxidize 5-methylcytosine (5mC) to 5-hydroxymethycytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), which may lead to eventual demethylation. In the genome, different oxidized products occupy overlapping, but largely distinct, genomic regions, and only a subset of 5hmC is further oxidized, which leads to eventual demethylation. However, how the distributions and the fates of oxidative derivatives of 5mC are determined in cells remains elusive.
Researchers from Prof. ZHU Bing’s group in the Institute of Biophysics (IBP) of the Chinese Academy of Sciences and their collaborators set up a DNA affinity purification-based semi-quantitative mass spectrometry strategy to identify proteins preferentially associating with 5hmC in vitro. They identified SALL4A as one of the top candidates that associates with 5hmC in vitro. To test whether SALL4A recognizes 5hmC in cells, they mapped the genome-wide distribution of SALL4A by chromatin immunoprecipitation followed by deep sequencing. In mouse embryonic stem cells, SALL4A localizes to enhancers, and its chromatin association is largely dependent on TET1. Surprisingly, when comparative analyzing the distribution of 5hmC in the genome, they found that 5hmC is greatly depleted at SALL4A-occupied enhancers, whereas further oxidation products, 5fC and 5caC, are highly enriched. Based on these results, the researchers conclude that most 5hmC at SALL4A-bound enhancers undergoes further oxidation.
Analysis of the DNA modification states in Sall4 knockout mouse embryonic stem cells reveal that the further oxidation process is abrogated upon deletion of Sall4 gene. TET1 and TET2 have similar biochemical activity, but the researchers found that TET2 is the main enzyme that oxidized 5hmC further at SALL4A-occupied enhancers. In collaboration with both TET1 and TET2, SALL4A induces 5hmC further oxidation at its associated enhancers by stabilizing the chromatin association of TET2. In this study, the researchers uncover an intriguing interplay between SALL4A and TET enzymes in oxidizing 5mC in a step-wise fashion. Such process thereby fine-tunes expression profiles of developmental genes in mouse embryonic stem cells and primes them for future expression during lineage commitment.
The research article, entitled “Cooperative Action between SALL4A and TET Proteins in Stepwise Oxidation of 5-Methylcytosine”, was published online in the journal Molecular Cell on November 10, 2016. This study is a collaboration work among the Institute of Biophysics (IBP), CAS; Tongji University; Research Center for Eco-Environmental Sciences, CAS; National Institute of Biological Sciences, Beijing; Kumamoto University and the Institute of Biochemistry and Cell Biology, Shanghai Institute for Biological Sciences, CAS. This work was supported by the China Natural Science Foundation, the Chinese Ministry of Science and Technology, the Strategic Priority Research Program of the Chinese Academy of Sciences, and the Howard Hughes Medical Institute International Early Career Scientist Program.
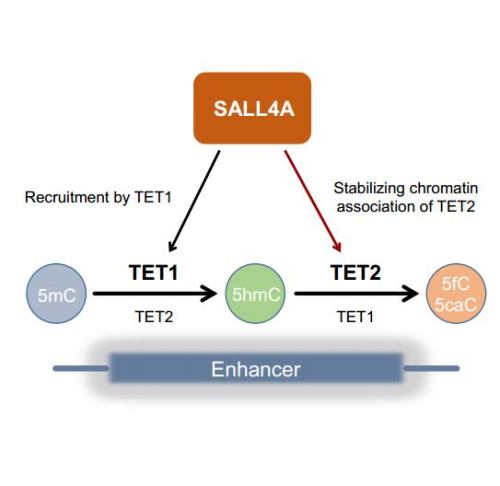
Figure: The graphic model of SALL4A facilitated stepwise 5mC oxidation by TET1 and TET2 (Image by IBP)
Contact
ZHU Bing
National Laboratory of Biomacromolecules, IBP
E-mail:zhubing@ibp.ac.cn
Tel:86-10-64888832

