Researchers at Institute of Biophysics Find New Rule Governing Box C/D RNA Recognition of Substrate
Box C/D RNAs are a large and ancient family of noncoding RNAs present in archaea and eukaryotes. They assemble with multiple proteins into ribonucleoprotein (RNP) enzymes that catalyze methylation of ribose at numerous sites in ribosomal RNAs and small nuclear RNAs. As specificity determinant, C/D RNAs contain one or two guide sequences that can form a stretch of base pairs with the substrate, thereby selecting a specific site for modification. The guide region is of variable length, particularly in eukaryotes, and is predicted to form 10-21 base pairs (bp) with substrates.
A research group led by Prof. YE Keqiong at the Institute of Biophysics of the Chinese Academy of Sciences, using a combination of biochemical and crystallographic analyses, found that box C/D guide RNAs can only form a duplex of a maximum of 10 bp with the substrate during modification. Longer duplexes must be unwound at one end in order to fit into a rigid and size-limiting protein channel for modification.
The same research group previously determined a C/D RNP structure that shows a 10-bp guide-substrate duplex capped at two ends by protein domains. To understand how longer duplexes are accommodated, a new structure was determined in which the guide can form an 11-bp duplex with the substrate by design. However, duplexes of only 10-bp long are formed, indicating that the protein channel is rather rigid and restraints the size of the duplex. In addition, elongated duplexes were found to inhibit the modification activity at low reaction temperature due to slow unwinding.
The finding provides new insight into how C/D RNAs recognize their substrates and how dimeric C/D RNPs are organized. Archaeal C/D RNPs were previously observed to form a dimeric species at in vitro reconstitution conditions. The structural organization and biological relevance of dimeric RNP are controversy. The C/D RNA has been proposed to be swapped between two different protein complexes in dimeric RNP. Identification of the 10-bp rule of substrate recognition is incompatible with the RNA-swapped model that lacks a size-limiting substrate-binding channel.
This study, entitled Box C/D guide RNAs recognize a maximum of 10 nt of substrates, was published online in the journal of Proceedings of the National Academy of Sciences of the United States of America on September 12, 2016. The work was supported by the National Natural Science Foundation of China, the Strategic Priority Research Program of the Chinese Academy of Sciences and the Beijing Municipal Government.
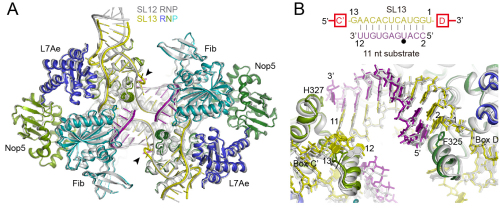
Structural alignment of SL13 and SL12 C/D RNPs (Image by YE Keqiong's group)
Contact person:
YE Keqiong
Key Laboratory of RNA Biology
Institute of Biophysics
Phone: 86-10-64887672
E-mail: yekeqiong@ibp.ac.cn

