Structural and Functional Analysis of Plant Photoprotective Protein PsbS
In romantic literature, every relationship even true love described is made up of love and hate without exception. This law also goes for nature. There is a love-hate relationship between plants and sunlight. On the one hand, it is absolutely necessary for plants to receive sunlight for their photosynthesis. On the other hand, with excessive sunlight, irreversible damage will be caused to the photosynthetic machinery of plants. To reduce or avoid such photooxidative damage, plants have evolved an efficient photoprotective mechanism called energy-dependent quenching (qE). Under high light conditions, the lumenal pH of plant thylakoid will drop from 6.5 to 5.5–5.8, leading to the activation of PsbS, a photosystem II protein embedded in the thylakoid membrane. The activated PsbS will initiate qE, enabling plants to safely dissipate the excessive absorbed light energy as heat.
The essential role of PsbS in qE was reported in 2000. However, its pigment-binding properties are still controversial, and its mechanism of action remains elusive. Therefore, solving the three-dimensional structure of PsbS is a long-awaited work in the photosynthesis field.Four years ago, Wenrui CHANG’s group from Institute of Biophysics, Chinese Academy of Sciences embarked on the structural investigation of PsbS to elucidate the mechanism of PsbS action in qE.
In a new study published online in Nature Structural & Molecular Biology in August 10, 2015, titled Crystal Structures of the PsbS Protein Essential for Photoprotection in Plants, Wenrui CHANG’s group released their structural and functional analysis of plant photoprotective protein PsbS.
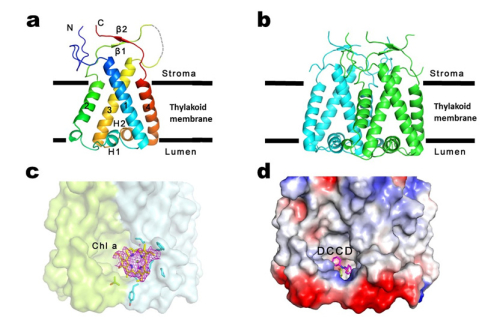
In their study, they developed a protocol via which a large quantity of homogeneous PsbS protein can be purified from spinach leaves. They purified and crystallized PsbS at low pH and solved its structure in the active state at 2.35 angstrom resolution. The crystal structure reveals that PsbS is composed of four transmembrane helices, forming a compact structure, which leads to no internal space left for formation of pigment-binding sites within PsbS monomer. The overall structure of PsbS is significantly different from those of LHCII and CP29, two members of the LHC superfamily whose structures have been solved by Chang’s group previously. Combining structural analyses and biochemical experiments, they demonstrated that PsbS is a compact dimer at low pH (in the active state). Moreover, based on the structural analysis of qE inhibitor DCCD-bound PsbS, together with biochemical experiments, they found that PsbS is a dimer in both inactive and active states, and that the transition between the two states involves the low pH-induced conformational change of the lumenal loops of PsbS dimer. Interestingly, they also found a chlorophyll a molecule at the PsbS dimer interface, which indicates that PsbS may participate in qE directly.
Their findings provide important insights into the mechanism of PsbS activation and inhibition and putative mechanism of PsbS action in qE. The reviewers of the manuscript spoke highly of the research and made the following comments: “the present work clearly represents an essential step forward”, “it represents a breakthrough in the understanding of the structural basis of photo protection in plants”, and “obtaining a structure of PsbS is a major breakthrough”.
Fig. Overall structures of PsbS. a. PsbS monomer; b. PsbS dimer; c. The potential chlorophyll a molecule at PsbS dimer interface; d. The qE inhibitor DCCD bound to PsbS dimer (image by Wenrui CHANG’s group )