Substrate-bound Structure of the E. coli Multidrug Resistance Transporter MdfA
Every coin has two sides. When people are infected with bacteria and suggested by doctors to take various drugs, the common issue they are worried about is if I take these drugs for a long period, whether the drug resistance will come into being?
Many bacteria pathogens have gained resistance to multiple types of antibiotics. As the greatest challenging threat to the public health all over the world, the research on the mechanism of bacteria drug resistance has grown to be a hot issue of common interest to WHO and governments worldwide.
According to previous findings, bacteria use various tactics to resist the toxic effects of antibiotics. One of the simple and effective ways is to pump drugs out of the cells using membrane-located transporters, similar to what cancer cells do to resist chemotherapy.
In a new study published in Cell Research on 4 August, 2015, titled “Substrate-bound Structure of the E. coli Multidrug Resistance Transporter MdfA”, Prof. ZHANG Kai’s group and Prof. ZHAO yongfang’s group released their structural and functional studies on a multidrug transporter from E. coli.
In their study, MdfA belongs to the largest secondary active transporter family, MFS. The crystal structure of MdfA-chloramphenicol complex at 2.4 angstrom resolution represents the first reported structure of an MFS multidrug transporter complexed with its substrate drug. MdfA forms 12 transmembrane helical topology structures, which contain two structural domains forming by 3 transmembrane helical repetitive structures. By means of mutation experiment, the group carried out research on the binding sites of chloramphenicol on the basis of its cell level, which identified amino acid residue who played an important role in several substrates combination and protonation loci. On the bases of this crystal structure and related biochemical-biophysical analyses, they proposed a general mechanism of the multidrug resistance by this type of transporters.
Their studies on these transporters will facilitate development of new strategies to overcome the drug-resistance problem and help us to understand better the nature of this endless war between human beings and bacteria.
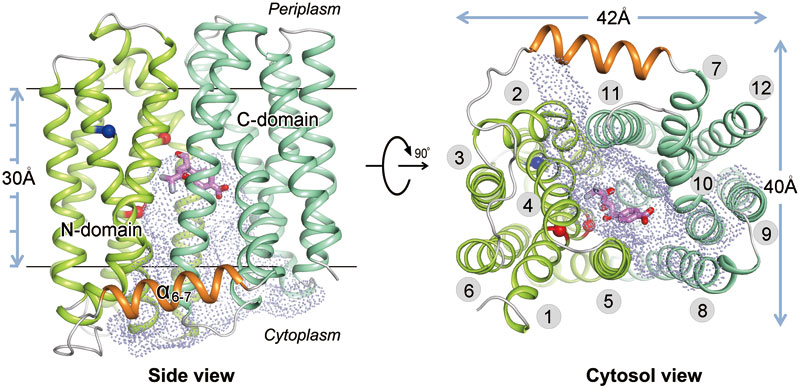
Fig. Overall structure of MdfA-Cm. The backbone of MdfA is shown in tube representation, with N-domain, C-domain and α6-7 in green, cyan and orange, respectively. The substrate Cm is shown as magenta stick model. Positions of E26 and D34 are marked as red spheres, and R112 as a blue sphere. The inward-facing cavity is shown as dot-surface representation. TMs are labelled in the right panel.(image by ZHANG Kai and ZHAO Yongfang, ? 2015 Shanghai Institutes for Biological Sciences)
CONTACT:
Prof. ZHANG Kai
Kai Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China.
E-mail: zhangc@ibp.ac.cn
Prof. ZHAO Yongfang
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China.
E-mail: yongfangzhao@ibp.ac.cn

