Novel molecular details of a complex mediating RNA-guided immune surveillance
The immune system protects an organism against a wide variety of pathogens. Clustered regularly interspaced short palindromic repeats (CRISPR) together with CRISPR-associated (Cas) proteins form the CRISPR/Cas system to defend against foreign nucleic acids.
Functions of biomolecules are intensively influenced by their structure. High resolution crystal structure reveals critical insight for understanding complex biological mechanisms.
Many studies had been conducted to determine the structure of CRISPR/Cas system. But due to its complexity, previous effort only obtained low-resolution cryo-electron microscopy structures, which provided limited information to explain the system’s function and mechanism.
Professor WANG Yanli at the Institute of Biophysics, Chinese Academy of Sciences and her colleagues recently reported a breakthrough in this field in the top-tier scientific journal Nature.
WANG’s team used Escherichia coli as model organism and studied the I–E subtype CRISPR/Cas system, which includes eleven subunits from five Cas proteins (CasA1B2C6D1E1) assemble along a CRISPR RNA (crRNA). They determined the 3.05 ? crystal structure of the 405-kilodalton Escherichia coli Cascade complex. (Fig 1)
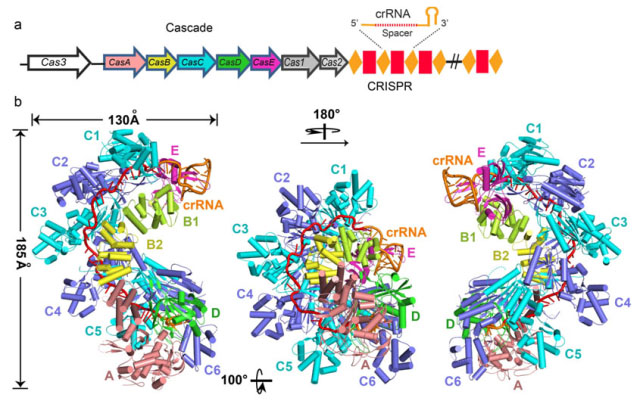
Fig 1. The 3.05 ? crystal structure of the 405-kilodalton Escherichia coli Cascade complex.
They found that the bound 61-nucleotide crRNA spans the entire 11-protein subunit-containing complex, where it interacts with all six CasC subunits (named CasC1–6), with its 5′ and 3′ terminal repeats anchored by CasD and CasE, respectively. The crRNA spacer region is positioned along a continuous groove on the concave surface generated by the aligned CasC1–6 subunits. The five long β-hairpins that project from individual CasC2–6 subunits extend across the crRNA, with each β-hairpin inserting into the gap between the last stacked base and its adjacent splayed counterpart, and positioned within the groove of the preceding CasC subunit. Therefore, instead of continuously stacking, the crRNA spacer region is divided into five equal fragments, with each fragment containing five stacked bases flanked by one flipped-out base. Each of those crRNA spacer fragments interacts with CasC in a similar fashion. Furthermore, their structure explains why the seed sequence, with its outwards-directed bases, has a critical role in target DNA recognition.
This work provides novel molecular details of protein–protein and protein–RNA alignments and interactions required for generation of a complex mediating RNA-guided immune surveillance.

