Molecular mechanism on the formation of R-loop structure in CRISPR-Cas9 system
CRISPR-Cas system confers adaptive immunity against exogenic elements in many bacteria and archaea. It uses small non-coding RNAs which are transcribed from invasive genetic elements as guide to target and cleave the invasive DNA. Recently, it have attracted much attentions due to the discovery that CRISPR-Cas9, a class II CRISPR-Cas system, can be re-purposed as a powerful RNA-guidedDNA targeting platform to easily create deletions, insertions and replacements in mammalian genome. Despite the wide usage of the powerful CRISPR-Cas9system in genome editing, numerous studies have assessed off-target DNA binding and cleavage by the Cas9–RNAcomplex, both in vitro and in vivo. Efforts to understand the molecular mechanism of DNA targeting and cleavage of Cas9,and improve its specificity and efficiency have accordingly been of great interests in this area.
Professor Jizhong Lou of the Institute of Biophysics (IBP) of the Chinese Academy of Sciences and colleagues examine the DNA cleavage activities of Streptococcus pyogenes Cas9 (SpyCas9) and its mutants using various target sequences and study the conformational dynamics of R-loop structure during target binding using single-molecule fluorescence energy transfer (smFRET) technique. They found that Cas9–sgRNA complex divides the target DNA into several distinct domains: proto-spacer adjacent motif, linker, Seed, Middle and Tail. After seed pairing, the Cas9 transiently retains a semi-active conformation and induces the cleavage of either target or non-target strand. smFRET studies demonstrate that an intermediate state exists in prior to the formation of the fully stable R-loop complex. Kinetics analysis of this new intermediate state indicates thatthe lifetime of this state increases when the base pairing length of guide-DNA hybrid duplex increasesand reaches the maximum at the size of 18 bp.
Based on their results and previous other work, they proposed an updated model for the formation of R-loop complex in SpyCas9 system (Figure 1). This model provides new insights into the process of R-loop formation and reveals the source of off-targeting in CRISPR/Cas9 system, and may shed new light on the sgRNA design and Cas9 engineering for optimizing CRISPR-Cas9 genome editing specificity.
The research work, entitled The initiation, propagation and dynamics of CRISPR-SpyCas9 R-loop complex, was published online in the journal Nucleic Acids Research on November 14, 2017. This work was supported by grants from the National Natural Science Foundation of China and the Ministry of Science and Technology 973 Project. Professor Yanli Wang provides insightful comments on the work.
Article linkage: https://academic.oup.com/nar/article/doi/10.1093/nar/gkx1117/4626768?guestAccessKey=ebef53b7-c7de-42a4-bb88-e2cc2397732e
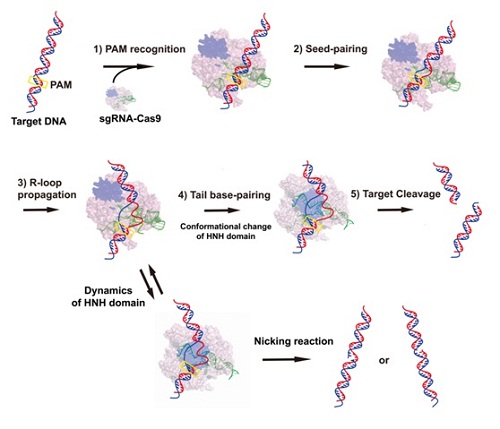
Figure 1. An updated model for the formation of R-loop complex in SpyCas9.

