Structural Basis for DNA Recognition by STAT6 Identified by IBP
Signal transducer and activator of transcription (STAT) family mediates transmission of signals of numerous cytokines and growth factors from the cell membrane to the nucleus via the classical JAK (Janus Activate Kinase)-STAT pathway. It has a significant role in allergic disease, asthma, immune responses and cancers and is of great importance in the development of therapeutic targets.
This study solved three crystal structures of STAT6 core fragment encompassing aa 123-658 in phosphorylated dimeric form and its complexes with N4 and N3 site DNA, respectively. They could thus, for the first time, analyze the conformational changes induced by DNA binding without having to resort to qualitative comparisons with phylogenetically distant homologues. A clear rotational conformational change was firstly observed upon DNA binding, and the conformation could be stabilized by DNA. Furthermore, they identified the key residues in the DNA binding domain that are important for DNA recognition and verified their roles in the function of STAT6 by performing in vitro and in vivo experiments. Notably, they showed that both a larger dimer interface intersection angle between the two protomers and the unique residue H415 in STAT6 DBD was important for N4 site DNA recognition (Fig. 1). This work is entitled Structural Basis for DNA recognition by STAT6 and published online by the Proc Natl Acad Sci USA on Nov 1, 2016.
A recent study reported that STAT6 plays a pivotal role in anti-viral signaling initiated by host cells in response to viral infections. Intriguingly, STAT6 could be activated by the STING/TBK1 cascade via phosphorylation of Y641. Surprisingly, this phosphorylation event occurred independently of the JAK family of kinases. Activation of STAT6 resulted in production of several types of chemokines that conferred protection against viral infections. Interestingly, residue S407 located in the DBD of STAT6 has been shown to be phosphorylated by TBK1. However, its implication for biological function of STAT6 is currently unknown. Thus, structural studies on STAT6 and its complex with DNA are essential to address several unanswered questions related to signals that morph STAT6 into a conformation, which is competent for binding DNA and transcription of genes.
STATs recognize DNA motifs with a consensus sequence of 5’-TTCN3/4GAA-3’ in the expression regulation regions of target genes, where N3/4 denotes a spacer consisting of 3 (N3) or 4 (N4) nucleotides of any of the four types found in the DNA. STATs differ from each other in their preferences for the length of the spacer between the two palindromic sites. STAT6 is reported as the only member that prefers N4 over N3 site DNA, consistent with existing experimental determination of STAT DNA-binding motifs. STAT5 is also reported weakly binds N4 site DNA. So far, only homodimeric structures of STAT1 and STAT3 in complex with N3 site DNA (phosphorylated STAT1 and STAT3, PDB_ID: 1BF5 and 1BGl, respectively; unphosphorylated STAT3, PDB_ID: 4E68) have been solved; and structures of unphosphorylated monomeric STAT1 (PDB_ID: 1YVL) and STAT3 (PDB_ID: 3CWG) and unphosphorylated dimeric STAT5α (PDB_ID: 1Y1U) in protein-alone form have been published. These structures shed light on the domain organization, nature of dimerization and mode of N3 site DNA binding. However, to the best of our knowledge, there was no instance where structures of a phosphorylated STAT protein alone and in complex with DNA were available for the same STAT protein. Over the past decades, this limits our understanding of the mechanisms underlying DNA recognition by STAT6 and the molecular basis for how STAT6 discriminates N4 site DNA from N3 type of DNA.
This work was supported by the Natural Science Foundation of China (grant nos. 31570875, 31330019, and 81590761), the Ministry of Science and Technology of China (grants nos. 2014CB910400, 2013CB911103), the Beijing Nova Program (grant no. Z141102001814020), and Youth Innovation Promotion Association CAS (Grant no. 2013065) to S.O.. The authors are grateful to the staff at Synchrotron Beamlines (17U1 and 19U of SSRF, 5.0.1 and 12.3.1 of Advanced Light Source, 23ID-C of GM/CA-CAT) for technical support; Y. Han, Y. Wang, P. Xue and Y. Y. Chen at the Protein Science Core Facility of IBP for technical help with initial X-Ray diffraction studies, automated protein crystallization, mass spectrometry and surface plasmon resonance assay experiments, respectively.
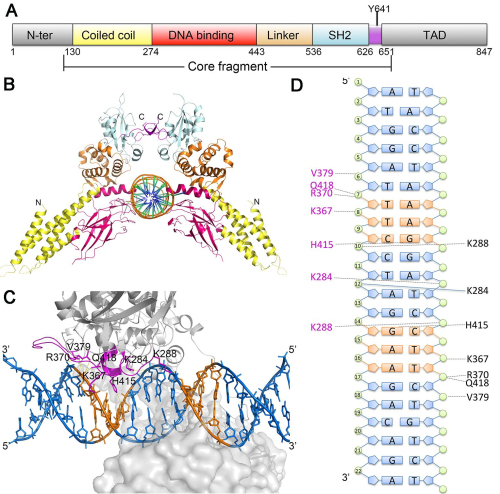
Fig.1 Structure of STAT6 and N4 site DNA complex.
Contact:
OUYANG Songying
National Laboratory of Biomacromolecules, IBP
E-mail:ouyangsy@ ibp.ac.cn
Tel:86-10-64888252

