WANG Xiaoqun's Group Generates an Atlas of Aging Human and Macaque Retina
The National Science Review journal published a research paper titled "A single-cell transcriptome atlas of the aging human and macaque retina" on August 26, 2020. With a focus on single-cell RNA sequencing of retina of humans and rhesus monkeys of different ages, the study uncovered the similarities and differences of cellular and molecular mechanisms between the two species, built a molecular atlas of the aging human and macaque retina, and found molecular changes and key molecular features of human retina in the aging process. It provided a potential intervention target to postpone retinal aging and a new thought to effectively prevent and treat age-related retinal diseases.
Vision is the most important sensory perception of both humans and animals. Over 80 percent of information has been received, handled and perceived by the visual system. Under the influence of light, photoreceptor cells of the retina will convert incoming light into electrochemical signals, which are then relayed by multiple neurons and eventually delivered by optical nerves to the brain for visual information. They are crucial for the survival of living organisms and will empower humans and animals to perceive the size, brightness, color and movement of objects. The function of retina suffers a decline and relevant patients are at high risk for retinal diseases in the process of aging. Therefore, it is of great importance for the prevention and treatment of age-related retinal diseases to understand molecular components and the changes of genetic regulatory network in retinal aging. As a special region in the retina of humans and non-human primate animals, macula lutea is primarily responsible for the perception of light and color. Many age-related eye diseases are caused by specific property of macula lutea. As the specialized fovea is absent in the retina of rodent models such as mice, thus the study of molecular mechanism of aging retina in humans and non-human primate animals is significant to treat age-related retinal diseases.
Researchers collected transcriptome atlas profiles of 119,520 single cells of the human and macaque retina from young to old stage in the study. With a comparison of cellular components and regional-specific molecular differences between human and macaque retina, they found that rods, which are generally believed to be homogenous, can be divided into MYO9A-positive and negative cells and that the proportion of two subtypes differs greatly in two species. Other types of cells in human and macaque retina demonstrated strong conservativeness in their gene expression model and cellular proportion. Furthermore, they conducted comparisons between foveal retina and peripheral retina. Their findings showed that Müller glia and cone cells have displayed obvious differences in foveal and peripheral gene expression and that the components of horizontal cells are diverse.
To investigate the molecular progression of aging human and macaque retina, researchers employed dataset of the aging to make a fitting of foveal and peripheral retinal figures and drew the aging trajectories in two regions. The degree of aging in macula lutea is higher than that in peripheral areas, which is identical with the neuroprotective effect of peripheral Müller glia gene expression. In addition, the study also found that rods suffer great damage in the process of aging, particularly a remarkable reduction of MYO9A-negative rods. Interestingly, the number of gene expression in foveal regions decrease greatly in the process of aging, while there is no noticeable reduction in the number of gene expression in peripheral areas. At last, researchers produced an atlas of gene expression associated with 55 types of human retinal diseases on the basis of retinal gene expression atlas of the two species and found the region-specific and cell-type gene expression. Nevertheless, there is no conservation in the gene expression model of some diseases between two species, indicating the importance of using human single-cell retinal atlas to study retinal diseases.
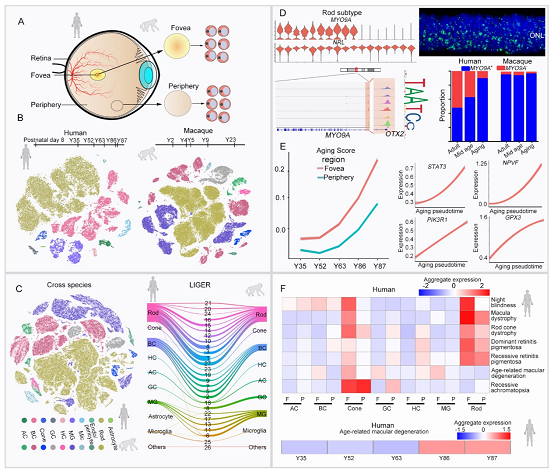
Figure: A single-cell transcriptome atlas of the aging human and macaque retina
(Image by Dr. WANG Xiaoqun's group)
Correspondence authors of the research article are Professor WANG Xiaoqun with the Institute of Biophysics (IBP) under the Chinese Academy of Sciences (CAS), Professor XUE Tian with the University of Science and Technology of China (USTC), Professor WU Qian with Beijing Normal University (BNU), and Deputy Researcher ZHANG Mei with the USTC. Doctoral Student YI Wenyang with the USTC, Doctoral Student LU Yufeng with the IBP, Postdoctoral Student ZHONG Suijuan with the BNU, and ZHANG are co-first authors of the article. It was funded by the Strategic Priority Research Program of the CAS, National Natural Science Foundation of China, National Key Research and Development Program of China, Beijing Municipal Science and Technology Commission, and Beijing Institute for Brain Research.
The web link for this research paper:
Contact: WANG Xiaoqun
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: xiaoqunwang@ibp.ac.cn
(Reported by Dr. WANG Xiaoqun's group)

