NyuWa Genome Resource of Variation Profile and Reference Panel for the Chinese Population Released
The lack of haplotype reference panels and whole-genome sequencing resources specific to the Chinese population has greatly hindered genetic studies in the world's largest population. Recently, groups of Prof. XU Tao and Prof. HE Shunmin from the Institute of Biophysics, Chinese Academy of Sciences reported the genome resource NyuWa (http://bigdata.ibp.ac.cn/NyuWa/) of Chinese genetic variation map and reference panel. This work was published in Cell Reports.
The Han Chinese population is the largest ethnic group in East Asia and even worldwide. Constructing an integrated, large-cohort, high-quality genetic variation database and reference panel for the Han Chinese population is imperative; such a resource would help clarify the population structure and population history, and facilitate genetic studies in the world's largest population.
In this work, the deep WGS data of 2,999 Chinese individuals were named as the mother goddess NyuWa, who was the creator of the human population in Chinese mythology. The NyuWa genome resource includes a total of 71.1M single nucleotide polymorphisms (SNPs) and 8.2M small insertions or deletions (InDels), including 25.0M novel variants and 22,504 potential loss-of-function variants in coding and noncoding genes.
According to annotation of clinical databases, there are 1140 pathogenic variants, and obvious differences of allele frequencies in known pharmacogenomic loci and cancer risk loci in different regions of China and populations worldwide. These results highlight the value of NyuWa genome resource in facilitating genetic and medical studies in Chinese and Asian populations.
Based on the NyuWa genome resource, a reference panel of 5,804 haplotypes and 19.3M variants was constructed, of which 3.25M specific variants are not included in other panels. Compared with other panels, the NyuWa reference panel reduces the Han Chinese imputation error rate by a margin ranging from 30% to 51%. NyuWa had an advantage over other panels in all allele frequency bins for the Chinese Han population. The imputation web server is publicly available on the NyuWa website.
In light of genetic differences between northern and southern Han Chinese people, samples from the NyuWa reference panel were divided into northern and southern subsets, and imputation performances were compared on independent public datasets. The results confirmed the applicability of one integrated panel for both northern and southern Chinese.
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences, the National Natural Science Foundation of China, the National Key R&D Program of China, the 13th Five-year Informatization Plan of Chinese Academy of Sciences, and the National Genomics Data Center, China.
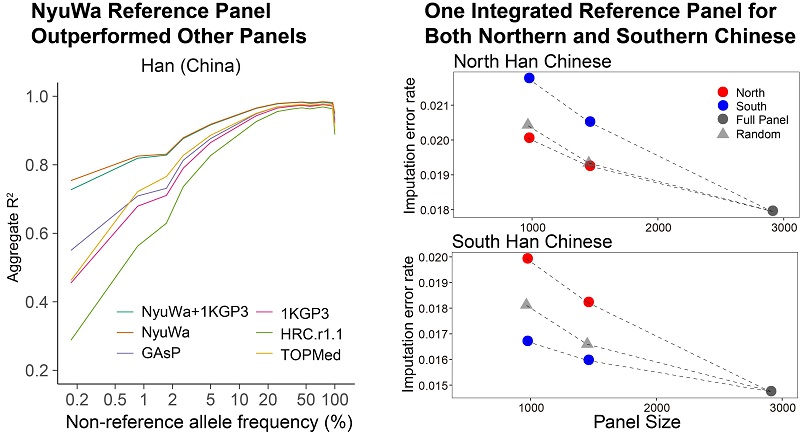
Imputation performance of NyuWa reference panel (Image by IBP)
Article link: https://www.sciencedirect.com/science/article/pii/S2211124721014996
Contact: HE Shunmin
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: heshunmin@ibp.ac.cn
(Reported by Dr. HE Shunmin's group)

