Cryo-ET Study from in vitro to in vivo reveals a general folding mode of chromatin
A research group led by Prof. ZHU Ping from the Institute of Biophysics of the Chinese Academy of Sciences has revealed a general folding mode of chromatin with two-start zigzag helical architecture both in vitro and in vivo, by using the novel cryo-electron tomography (cryo-ET) technology (Fig. 1). This new finding sheds light on the general architecture and folding mode of chromatin fiber, and the formation of condensed chromatin areas in vivo. The study was published in Cell Reports on Sept. 13, 2023.
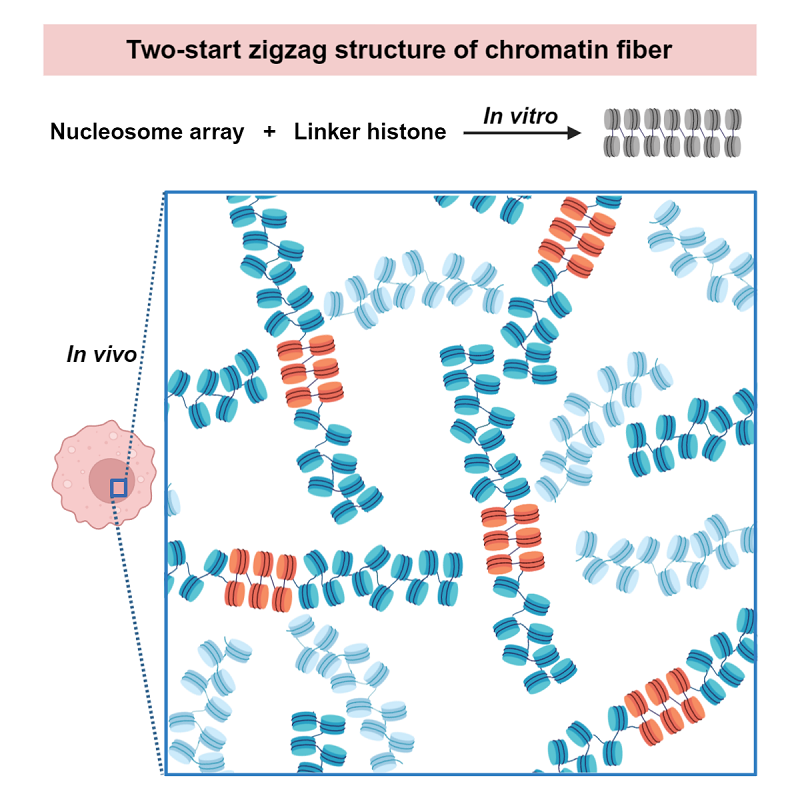
Fig 1. The double helical structure of chromatin with a general two-start zigzag folding mode
The organization and dynamics of chromatin fiber play crucial roles in regulating DNA accessibility for gene expression. Previous studies indicate that, in the presence of linker histones (or divalent cations), nucleosome arrays fold into a condensed chromatin fiber, which is considered as the platform for the gene transcription regulation. However, the structure and folding mode of chromatin, particularly in vivo, remains largely unknown.
In the present study, the researchers combine cryo-ET, sub-volume averaging, and 3D segmentation techniques to visualize the chromatin fibers folding by linker histone both in vitro and in vivo. In the in vitro system, they discover that an increased NRL and prolonged fiber length do not change the two-start helical architecture of the reconstituted long chromatin fibers.
Compared with the in vitro reconstitution system, the chromatin directly isolated from the HeLa cells, which contains native nucleosomes, appears to be more heterogeneous as it endures different NRL and DNA sequences, histone modifications, histone variants, etc, as is common in vivo. Nevertheless, researchers find that the isolated chromatin fiber also presents a two-start zigzag helical architecture, compatible with the in vitro reconstituted chromatin.
In the in vivo study, researchers analyze the structure and organization of chromatin inside the frog erythrocyte nuclei in situ by using a combining of technologies of cryo-focused ion beam milling (cryo-FIB), cryo-electron tomography and sub-tomogram averaging. In order to identify the spatial arrangement of nucleosomes in vivo, cryo-ET with voltage phase plate (VPP) is further used to directly visualize the DNA trajectory between the consecutive nucleosomes. According to their results, a two-start zigzag helical architecture of chromatin with unambiguous DNA trajectories between alternate N/N+2 nucleosomes is clearly determined inside the frog erythrocyte nuclei in situ.
Putting together, researchers propose a model that chromatin presents a general two-start zigzag folding mode which results a double helical fiber both in vitro and in vivo. "Our findings shed light on the general architecture and folding mode of chromatin fiber, and the formation of condensed chromatin areas in vivo," said Prof. Zhu.
Article link: https://www.cell.com/cell-reports/fulltext/S2211-1247(23)01146-4
Contact: ZHU Ping
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: zhup@ibp.ac.cn
(Reported by Prof. ZHU Ping's group)

