New Algorithm SIRM for Reducing Preferred Orientation Problem in Cryo-EM Three-dimensional Reconstruction
In a study published in Science Advances, a research team led by Prof. ZHANG Xinzheng from the Institute of Biophysics, Chinese Academy of Sciences, introduced the new algorithm "Signal-to-Noise Ratio Iterative Reconstruction Method (SIRM)", which purely utilizes a computational method to eliminate or reduce distortions in cryo-electron microscopy (cryo-EM) three-dimensional reconstructions caused by preferred orientation problem.
The cryo-sample preparation is a major bottleneck in single-particle analysis (SPA) techniques, with the issue of preferred orientation being one of the most common and challenging problems caused by cryo-sample preparation.
The SIRM method first removes misaligned particles from datasets with orientation bias (these misaligned particles stem from the preferred orientation problem); based on Fourier component signal-to-noise ratio, it performs dual-space-constrained iterative reconstruction in Fourier space and real space to correct distortions in three-dimensional reconstructions and reduce resolution anisotropy. Through data analysis, researchers found that this method can further enhance the accuracy of three-dimensional reconstructions in datasets with preferred orientation, thereby improving the resolution of three-dimensional reconstructions.
Using the SIRM method, researchers processed several datasets with preferred orientation. The results showed a significant improvement in resolution in the direction of the missing region in the reconstructed images after using the SIRM method. It effectively restored density distortions and resulted in resolution enhancement.
Compared to experimental methods, this approach relies solely on computational algorithms, offering advantages of low cost, ease of operation, and clear principles. It holds great potential in obtaining high-resolution three-dimensional structures of important protein complexes.
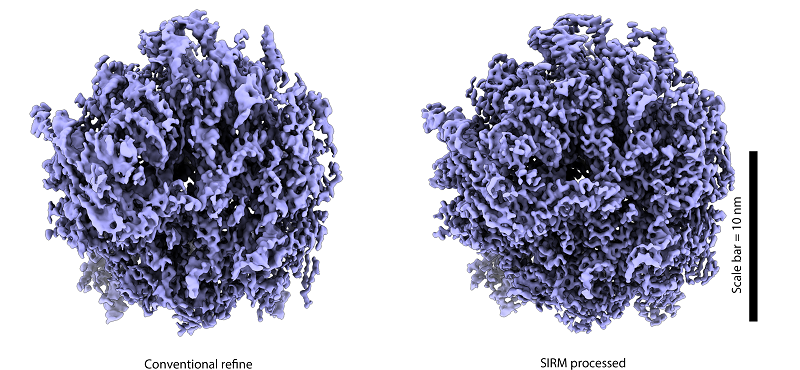
Fig. Comparison before and after applying the SIRM algorithm on ribosome datasets
(Image by ZHANG Xinzheng's group)
Article link: https://www.science.org/doi/10.1126/sciadv.adn0092
Contact: ZHANG Xinzheng
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
E-mail: xzzhang@ibp.ac.cn
(Reported by Prof. ZHANG Xinzheng's group)

