Scientists Reveal Advanced Structural Formation Mechanism of Nucleosome Binding and Chromatin Folding Mediated by Linker Histone H5
Recently, the research groups of Prof. ZHU Ping and Prof. LI Guohong from the Institute of Biophysics, Chinese Academy of Sciences, after long-term efforts, obtained the cryo-electron microscopy structure of chromatin particles formed by the folding of dodecanucleosome mediated by the linker histone H5 at a resolution of 3.6 angstroms (Å). This study reported, for the first time, the complete chromatosome structure formed by the full-length linker histone H5 (including its N-terminal and C-terminal domains) interacting with the nucleosome core particles (NCP), and established the full atomic structure model of the 30-nm chromatin fiber mediated by linker histone H5.
This study was published in Cell Research on August 5, 2024.
In eukaryotes, the folding pattern and regulatory mechanisms of the 30-nm chromatin fiber connecting the nucleosome beads and higher-order chromatin structures are crucial for understanding chromatin structure and its biological significance in regulating gene expression and other DNA-dependent activities.
In this study, scientists unraveled the cryo-EM structure of the H5-bound dodecanucleosome chromatin (referred to as H5-chromatin fiber) assembled from 12×177bp_601 DNA, core histones (H2A, H2B, H3, and H4), and linker histone H5 at a resolution of 3.6 Å.
The researchers found that H5-30-nm chromatin fibers form a structural unit with four nucleosomes, twisted and folded in a Zig-Zag manner between units, creating a left-handed double helix structure resembling a right-handed double helix with DNA. Based on the high-resolution cryo-EM density map obtained in this study, researchers established a complete atomic structure model of the H5-30-nm chromatin fiber.
In this research, scientists obtained the full-length structure of linker histone H5, including its N-terminal domain (NTD) and C-terminal domain (CTD) at atomic resolution, establishing for the first time an atomic resolution structure of a complete chromatosome (nucleosome core particle NCP + full-length linker histone).
Researchers found that the formation of the higher-order structure of the chromatin fiber exhibits structural asymmetry. By utilizing a series of genetics, genomics, and biophysics methods, researchers systematically validated the physiological functions and structural characteristics of the tetranucleosome folding unit in the budding yeast eukaryote. Based on these results, researchers proposed a molecular mechanism model for nucleosome binding and chromatin folding mediated by linker histone H5 and the formation of higher-order structures.
The results of this study are significant for establishing a fine model of high-resolution chromatin fiber structure, as well as for research on the assembly of higher-order chromatin structure and the epigenetic regulatory mechanisms, providing an important structural basis for understanding the molecular foundation of in vivo chromatin structure establishment and the potential mechanisms by which various epigenetic factors regulate chromatin structure.
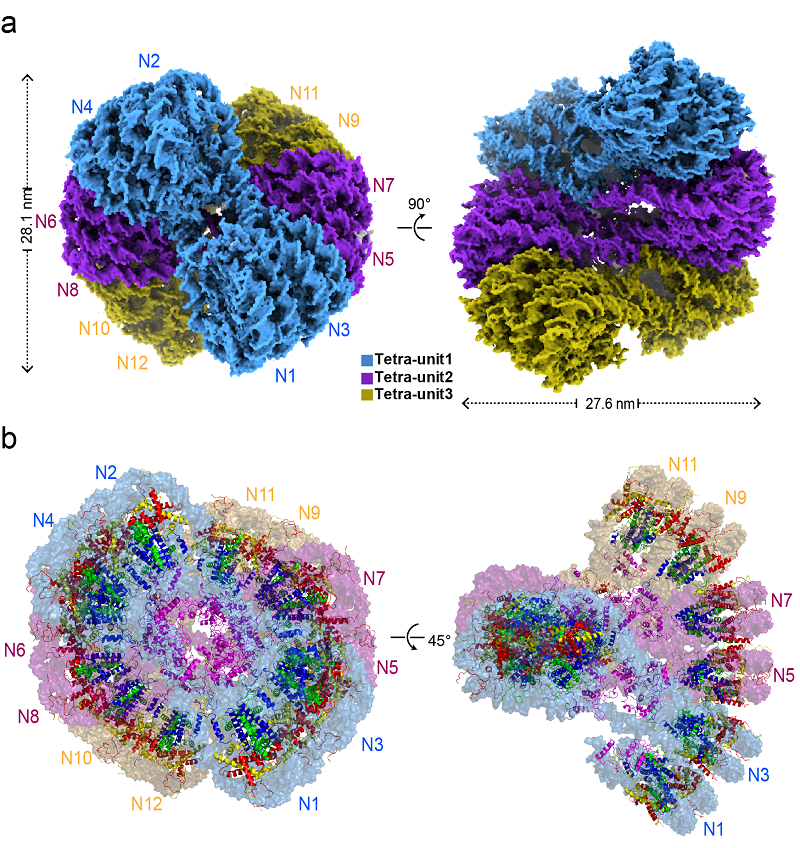
Figure 1. Cryo-EM structure of H5-30-nm chromatin fiber (a) and atomic structure model (b)
(Image by ZHU Ping's group)
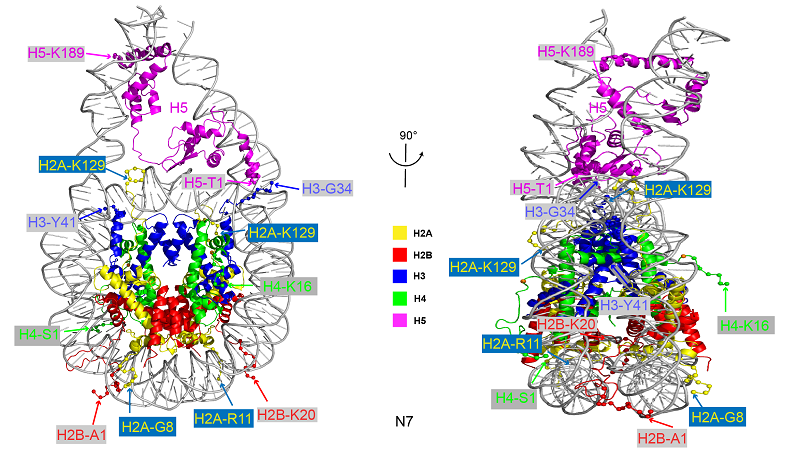
Figure 2. Complete chromatosome structure formed by full-length linker histone H5 and nucleosome core particle
(Image by ZHU Ping's group)
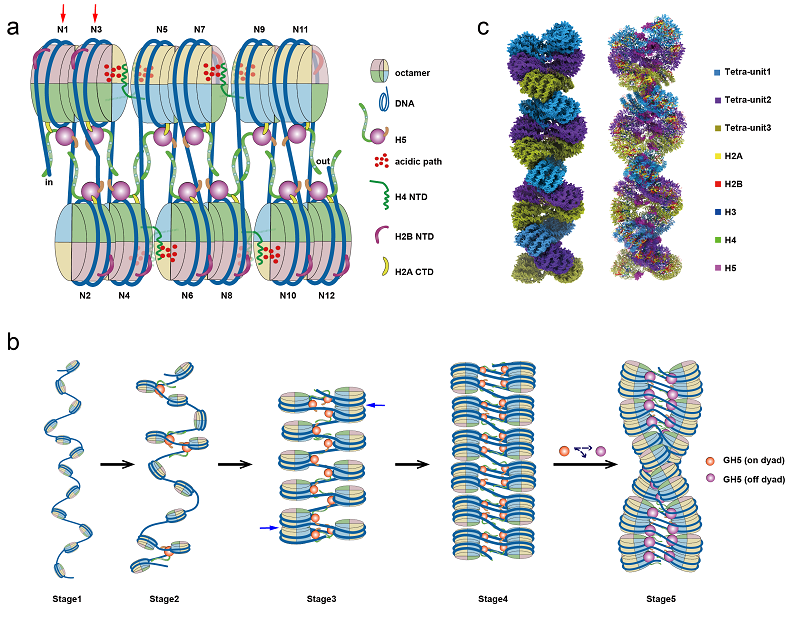
Figure 3. Model of linker histone-mediated folding and higher-order structural assembly of the 30-nm chromatin fiber
(Image by ZHU Ping's group)
Article link: https://www.nature.com/articles/s41422-024-01009-z
Contact: ZHU Ping
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: zhup@ibp.ac.cn
(Reported by Prof. ZHU Ping's group)

