Researchers Decipher the Polymorphism Map of Human Genomic Minisatellites
In the human genome, more than half of the regions are composed of repetitive sequences. These repetitive sequences are highly variable during human evolution, greatly enriching the genetic diversity of the genome. Variable number tandem repeats (VNTRs), also known as "minisatellites," are a type of DNA tandem repeat where the motif (repeat unit) is longer than 6 base pairs (bp), and the repeat number of motifs typically ranges from 10 to 60. Studies have shown that VNTRs can not only increase disease risk through length expansion but also have independent pathogenic effects due to changes in their motifs. However, due to limitations in sample size, sequencing depth, population diversity, and identification algorithms, genetic research on VNTRs in existing large-scale genomic sequencing projects is far from sufficient, leading to a portion of "missing" heritability in the human genome.
Recently, a research team led by Prof. XU Tao and Prof. HE Shunmin from the Institute of Biophysics, Chinese Academy of Sciences, constructed a global population VNTR polymorphism genetic map (NyuWa VNTR polymorphism map), systematically analyzed the functional characteristics of VNTRs, especially their regulatory roles in gene expression, using high-depth whole-genome sequencing data from 8,222 individuals across more than 140 countries and regions. Their findings were published in Cell Genomics.
Previously, NyuWa Genome Project, based on large-scale population whole-genome sequencing data, has published and analyzed human genomic SNP, InDel variation maps (Cell Reports, 2021), mobile element variation maps (MEI, Nucleic Acids Research, 2022), microsatellite variation maps (STR, Nature Communications, 2023) and has also explored the impact of recent positive selection in the genome (Science Bulletin, 2023) as well as adaptive selection of non-coding regulatory elements (Molecular Biology and Evolution, 2024) on human phenotypes and disease evolution.
The newly released NyuWa VNTR polymorphism map completes the NyuWa genomic resources' comprehensive coverage of the main repetitive elements in the human genome (ME, STR, and VNTR), serving as an important reference for systematically analyzing repetitive sequence element variations.
This work expands the scale of repetitive element variations in existing genetic research, providing a new perspective for understanding the role of repetitive elements in gene regulation, and offering significant references for future clinical research and genotype-phenotype association studies.
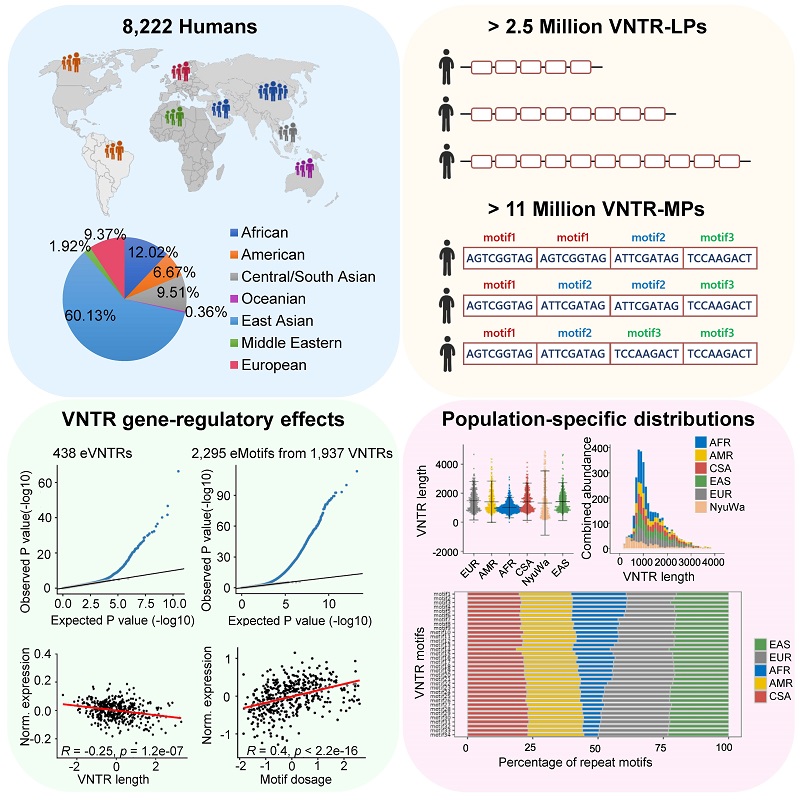
Fig. Graphical abstract of the article
(Image by HE Shunmin's group)
Article link: https://www.cell.com/cell-genomics/fulltext/S2666-979X(24)00328-8
Contact: HE Shunmin
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: heshunmin@ibp.ac.cn
(Reported by Prof. HE Shunmin's group)

