Compact Cas9d Enzyme Revealed as Promising Genome-Editing Tool
Cas9 is the hallmark protein of Type II CRISPR-Cas systems. Among them, Streptococcus pyogenes Cas9 (SpCas9) has become the most widely used Cas9 due to its high cleavage efficiency and robust genome-editing performance. However, its large molecular weight limits delivery through adeno-associated virus (AAV) vectors. Therefore, identifying naturally occurring or engineered Cas9 variants with smaller molecular size yet comparable cleavage activity has become a key need in the field of genome editing.
On August 11, 2025, a research team led by Prof. WANG Yanli at the Institute of Biophysics of Chinese Academy of Sciences, published a paper in Nature Communications reporting the structure and mechanism of a highly active Type II-D Cas9.
The study focused on a Type II-D Cas9 derived from a Nitrospirae bacterium (NsCas9d), which consists of only 762 amino acids. From metagenomic datasets, the researchers identified the CRISPR repeats and spacers associated with NsCas9d and designed a corresponding sgRNA.
In vitro cleavage assays showed that NsCas9d requires at least a 20-nt pairing between the substrate dsDNA and sgRNA to achieve robust dsDNA cleavage activity comparable to SpCas9.
Through PAM depletion assays combined with high-throughput sequencing, the researchers identified that NsCas9d recognizes a 5′-NRG-3′ PAM sequence, with the 5′-NGG-3′ PAM exhibiting the highest cleavage efficiency in vitro.
The researchers further resolved the cryo-EM structure of the NsCas9d-sgRNA-dsDNA ternary complex at 2.86 Å resolution, marking the first report of a complete HNH and RuvC domain structure for a Type II-D Cas9.
In the structural model, the dsDNA PAM sequence is accommodated within a positively charged binding channel formed by the PI and WED domains. Importantly, NsCas9d generates 3-nt 5′ overhangs as cleavage products, a sticky-end feature that can improve the efficiency and predictability of DNA repair processes during gene editing operations such as insertions.
This study not only deepens the understanding of Cas9 evolution and molecular mechanisms but also highlights the potential of NsCas9d as a compact and efficient tool for genome editing applications.
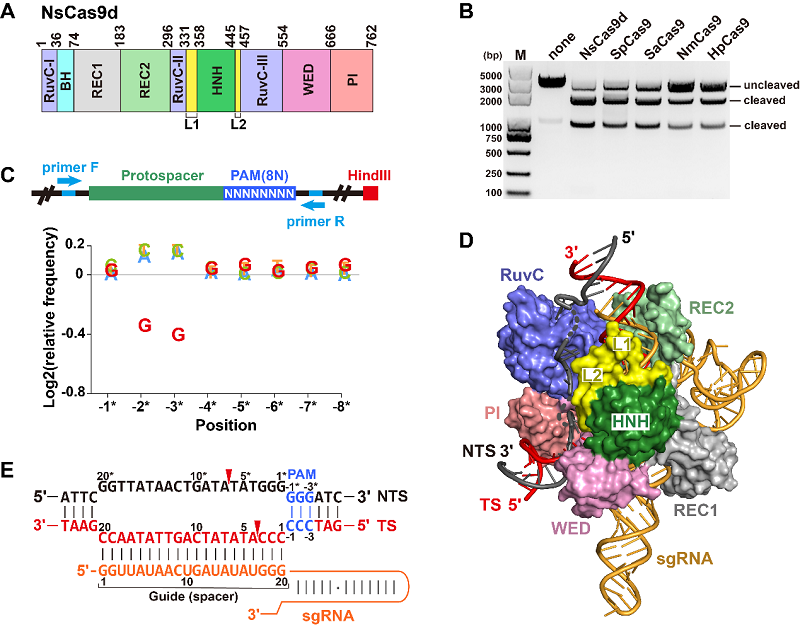
Figure: Cleavage activity of NsCas9d and structural model of the ternary complex
(Image by WANG Yanli's group)
Article link: https://doi.org/10.1038/s41467-025-62128-8
Contact: WANG Yanli
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
E-mail: ylwang@ibp.ac.cn
(Reported by Prof. WANG Yanli's group)

