Histone H3.3 Phosphorylation Regulates Chromatin Dynamics
The serine 31 (Ser31) phosphorylation modification (H3.3S31ph) in the N-terminal tail of the histone variant H3.3 can transform H3.3 nucleosomes from a stable state to a dynamically activated configuration, providing new mechanistic insights into epigenetic regulation. However, the specific mechanisms by which H3.3 influences nucleosome stability and dynamics remain unclear.
On September 12, 2025, a research team led by Prof. LI Wei from the Institute of Biophysics of the Chinese Academy of Sciences, and Prof. CHEN Ping from the Capital Medical University published a study in Nucleic Acids Research.
The study provides novel mechanistic insights for the first time into how H3.3 and its Ser31 phosphorylation regulate nucleosome dynamics and transcriptional responses, highlighting the critical role of this mechanism in macrophage immune response pathways.
Through a combination of in vitro single-molecule experiments and genome-wide analyses, this study demonstrates that while the incorporation of H3.3 does not significantly alter the mechanical stability of nucleosomes, it markedly enhances their ability to maintain integrity after disruption.
A key finding is that H3.3 recruits the FACT complex (Facilitates Chromatin Transcription) more efficiently than canonical H3. FACT typically destabilizes nucleosomes, but when bound to H3.3-containing nucleosomes, it instead promotes a stable, maintenance-oriented state.
The research further identifies phosphorylation at Ser31 of H3.3 as a critical molecular switch that reverses this stability. H3.3S31ph transforms the nucleosome from a stable state to a dynamic, active configuration, facilitating rapid transcriptional activation.
This modification is specifically induced in macrophages upon stimulation, where it dynamically modulates FACT binding, nucleosome states, and the ensuing transcriptional response.
These findings provide novel mechanistic insights into how the interplay between a histone variant, its post-translational modification, and a histone chaperone complex can precisely regulate chromatin dynamics.
The study underscores the importance of H3.3S31ph as a pivotal regulator enabling a rapid shift from transcriptional poising to activation in immune responses. This work also suggests potential therapeutic implications for diseases involving chromatin dysregulation, such as inflammatory disorders and cancer.
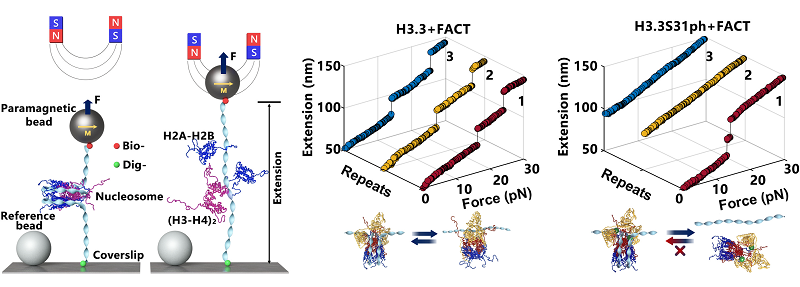
Figure 1. Precise characterization of the interaction between FACT and H3.3 nucleosomes using high-resolution single-molecule magnetic tweezers manipulation technology.
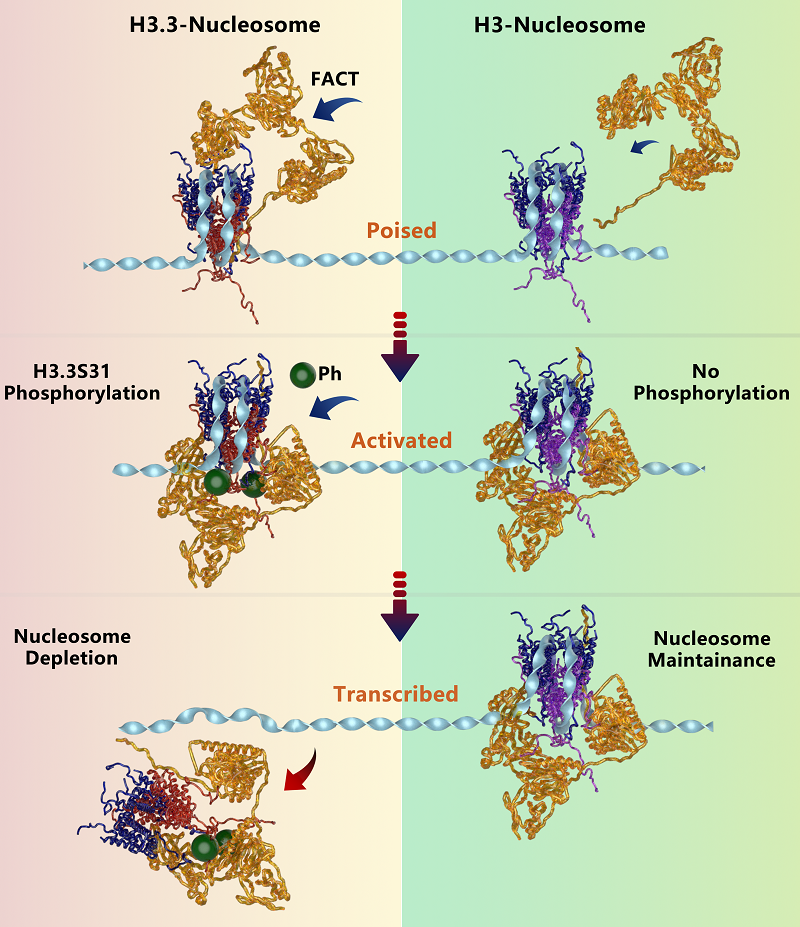
Figure 2. H3.3S31ph acts as a molecular switch regulating the dynamic transition of nucleosomes from a stable to an activated state.
(Image by LI Wei's group)
Article link: https://academic.oup.com/nar/article/53/17/gkaf891/8252030
Contact: LI Wei
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
E-mail: weili007@ibp.ac.cn
(Reported by Prof. LI Wei's group)

