Scientists Develop CRISPR PRO-LiveFISH for Live-Cell Genome Imaging
Existing CRISPR-Cas-based imaging methods can target endogenous genomic sequences, but their applications are limited by system complexity and sensitivity-particularly in imaging non-repetitive loci, performing multi-locus visualization, and working with primary cells.
To address these challenges, a joint research team from the Institute of Biophysics of the Chinese Academy of Sciences, and Tsinghua University has developed a novel live-cell DNA imaging technology called CRISPR PRO-LiveFISH (Pooled gRNAs with Orthogonal bases LiveFISH), which integrates CRISPR technology with expanded genetic alphabet technology.
Building upon the previous CRISPR LiveFISH approach, this upgraded system introduces, for the first time, orthogonal unnatural base pairs from an expanded genetic alphabet for site-specific fluorescent labeling of single guide RNAs (sgRNAs). Through rational design of Cas9-sgRNA structures, the team achieved a substantial improvement in probe labeling sensitivity.
This research was published in Nature Biotechnology on November 6, 2025.
By integrating DNA library construction with expanded genetic code technology, the PRO-LiveFISH method combines in vitro transcription and post-transcriptional modification strategies to generate a high-throughput, site-specific, and efficiently labeled sgRNA probe library. This enables high-sensitivity live-cell imaging of non-repetitive DNA sequences, including in primary cells. The method not only simplifies experimental procedures but also minimizes potential interference with native genomic structure and function.
Using PRO-LiveFISH, the researchers achieved simultaneous real-time tracking of multiple genomic loci within living cells. They systematically uncovered the relationship between chromatin dynamics and epigenetic states, elucidated the spatiotemporal characteristics of enhancer-promoter (E-P) interactions, and revealed the critical role of the transcriptional coactivator BRD4 in maintaining super-enhancer three-dimensional (3D) interactions. These findings deepen our understanding of epigenetic regulation and the spatiotemporal dynamics of the 3D genome.
With its high sensitivity, specificity, and compatibility with multiple cell types, CRISPR PRO-LiveFISH opens new avenues for exploring how 3D genome organization and epigenetic modifications coordinate to control gene expression, cell identity, and disease processes such as cancer.
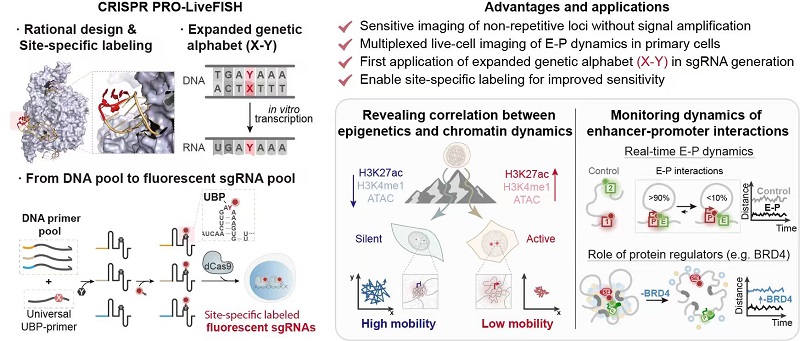
Figure: Design Strategy and Applications of CRISPR PRO-LiveFISH
(Image by FANG Xianyang's group)
Article link: https://www.nature.com/articles/s41587-025-02887-3
Contact: FANG Xianyang
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
E-mail: fangxy@ibp.ac.cn
(Reported by Prof. FANG Xianyang's group)

