Researchers developed molecular-scale 3D isotropic nanoscopy
Single-molecule localization microscopy (SMLM) has revolutionized biological imaging by enabling nanoscale visualization of cellular structures. However, its practical application has been hampered by one persistent challenge: significantly lower axial resolution compared to lateral resolution, resulting in anisotropic blurring that distorts three-dimensional (3D) reconstructions.
On 2 December 2025 , a team of researchers led by Prof. XU Tao and Prof. JI Wei at the Institute of Biophysics (IBP) of the Chinese Academy of Sciences (CAS) developed a technique named repetitive optical selective exposure in 3D (ROSE-3D), which achieves isotropic nanoscale resolution in 3D SMLM by incorporating interferometric illumination along the X, Y and Z axes.
For in-focus molecules, ROSE-3D enhances lateral localization precision by a factor of two and axial precision by 3.5 times compared to conventional astigmatic approaches. Importantly, under defocused conditions, which typically degrade image quality, ROSE-3D's minimal sensitivity to defocus-induced distortions leads to improvements of 6-fold in lateral and 8-fold in axial localization precision.
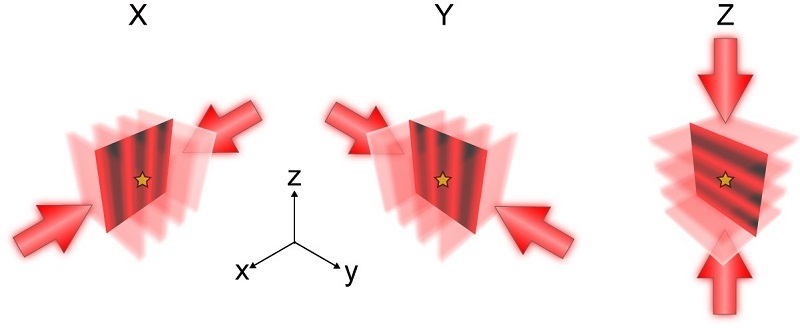
Figure 1. Principle of ROSE-3D.
(Image by XU Tao and JI Wei's group)
Leveraging the 3D resolving power of ROSE-3D, the team resolved the hollow structure of microtubules across the entire 1-micrometer thickness, a challenging achievement not previously attainable with standard methods.
Extending ROSE-3D to multicolor and multilayer imaging, the researchers performed two-color imaging of lamin A/C and lamin B1 across the entire nucleus of COS-7 cells. Statistical analysis revealed that lamin A/C is distributed inside lamin B1 with a spacing of approximately 10 nm. This marks the first time such a distribution has been observed from a 3D whole-nucleus perspective.
Moreover, utilizing the isotropic 3D resolution capabilities of ROSE-3D, the team visualized the mitochondrial fission protein DRP1 and its associated assemblies on the mitochondrial outer membrane. These assemblies were classified into four distinct morphological types, including helical and ring-like structures, providing the first in situ morphological characterization of DRP1 complexes within cells.
These results demonstrate the outstanding 3D resolution of ROSE-3D, along with its robust multicolor and multilayer imaging capabilities, positioning it as a powerful tool for investigating 3D nanoscale architectures within cells.
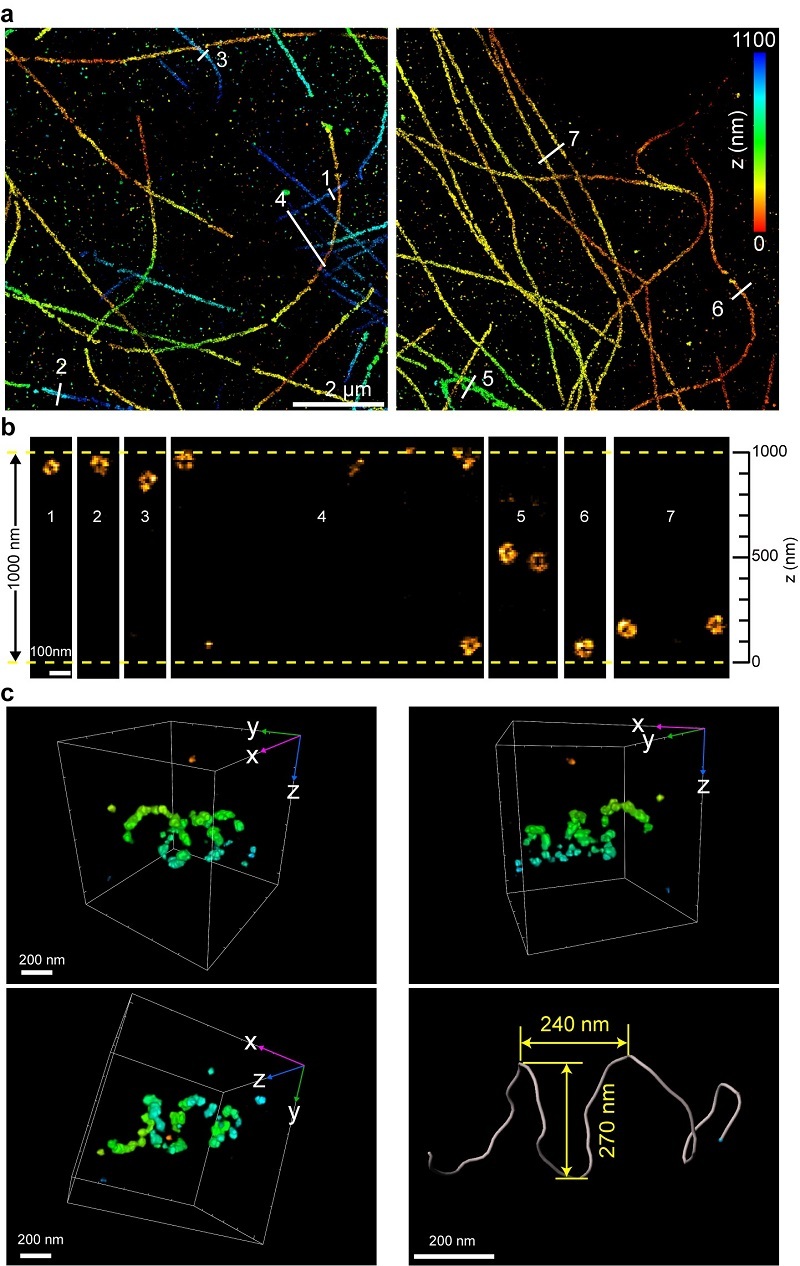
Figure 2. ROSE-3D resolved the hollow structure of microtubules (a-b) and the helical structure of DRP1 complexes (c).
(Image by XU Tao and JI Wei's group)
Article link: https://www.nature.com/articles/s41592-025-02911-z
Contact: XU Tao, JI Wei
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
E-mail: xutao@ibp.ac.cn, jiwei@ibp.ac.cn
(Reported by Prof. XU Tao and Prof. JI Wei's group)

