Research unveils histone H3K27 acetylation is dispensable for enhancer activity in mouse ES cells
Enhancers are key regulatory elements that control the spatial and temporal gene expression, which is vital for cell differentiation and disease occurrence. It has been an important research focus to precisely identify DNA sequences that execute enhancer functions in the genome. In the post-genomics era, with the assistance of next-generation sequencing, researchers began to utilize the numerous signatures of chromatin, including DNA methylation, histone modifications, chromatin accessibility, and the distribution of chromatin proteins, to annotate important elements in the genome. Among these signatures, H3K4me1, H3K27ac, highly enriched histone variant H3.3, and a high histone turnover rate are remarkable markers foractive enhancers. However, whether H3K27ac, the best indicator of active enhancers, could functionally determinate enhancer activity is less clear.
A new study published online in Genome Biology on February 21, 2020, titled "Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells", reports that selective erasure of H3K27ac at enhancers do not impact active transcription. Thus, enhancer activity does not solely depend on H3K27ac, instead it is a functional consequence of the synergistic action by multiple acetylation events at different histone sites.
To specifically target enhancer H3K27ac, the researchers mutated histone variant lysine 27 in H3.3 to arginine (H3.3K27R) and produced non-acetyl-modifiable H3.3 histones in mouse ESCs. H3K27ac is almost completely depleted at active enhancers in H3.3K27R mutant ESCs. Meanwhile, the transcriptome change is minimal. The authors applied the newly developed model, activity-by-contact, to define target genes of H3K27ac-marked active enhancers, and found that the transcription of these genes is not sensitive to the dramatic decrease in H3K27ac at their enhancers. Transcription of genes associated with super enhancers, which harbor enormous amounts of H3K27ac, is also mostly unchanged.
The authors further profiled the distributions of acetylation at other histone lysine residues, including H3K9, H3K18, H3K122, H4K5, and H4K8, and found that acetylationsat these sites are remained genome-widely. Acetylation can neutralize the positive charge on lysine residues, leading to attenuated interaction between histone tails and DNA. Therefore, it is the synergistic effect of multiple acetylation events that crucial for chromatin opening and maintaining enhancer activity. This study clearly showed that although H3K27ac is, and remains to be the best indicator of active enhancers in the mammalian genome, its depletion at enhancer regions does not affect chromatin accessibility and gene transcription. This study suggests that H3K27ac has to work in concert with acetylation events on other histone lysine residues.
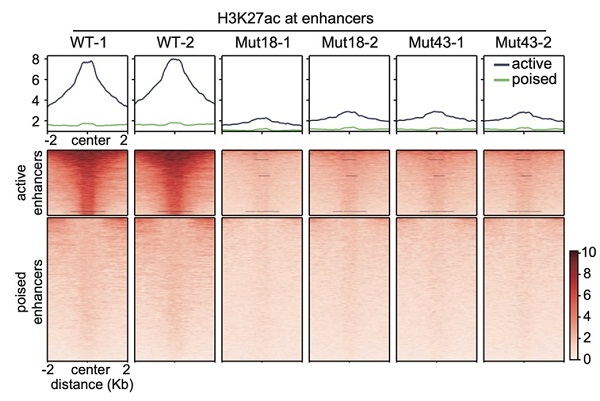
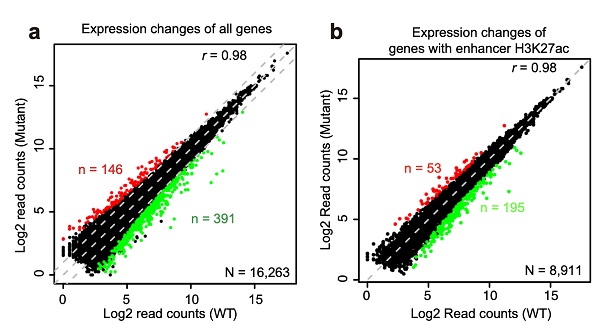
Depletion of Enhancer H3K27ac does not Affect Transcription
(Image by Dr. ZHU Bing's group)
This work is achieved by ZHU Bing laboratory, Institute of Biophysics, Chinese Academy of Sciences. Institute of Biophysics and University of Chinese Academy of Sciences are the first and second institutions, respectively. Prof. ZHU Bing and Dr. XIONG Jun are the corresponding authors. ZHANG Tiantian and Dr. ZHANG Zhuqiang are the co-first authors of this paper. Dr. DONG Qiang also contributed to this study. This work is supported by the Ministry of Science and Technology, the Chinese Academy of Sciences, and Natural Science Foundation.
Article link: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-020-01957-w
Contact: Bing Zhu
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101,China
Phone: 86-10-64888832
Email: zhubing@ibp.ac.cn
(Reported by Dr. ZHU Bing's group)

