Chinese scientists reveal the molecular mechanism of pyroptosis executioner GSDMD recognition by activated caspases in innate immunity
On Feb 27, 2020, a research article entitled "Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis" was published in Cell by a collaborative study of WANG Dacheng -DING Jingjin's group from the Institute of Biophysics, Chinese Academy of Sciences, and SHAO Feng's group from National Institute of Biological Sciences, Beijing. They deciphered the precise molecular mechanism of pyroptosis executioner GSDMD recognition by activated caspases in innate immunity.
Pyroptosis is one of the most important innate defenses against infections and endogenous dangers. Excessive pyroptosis causes inflammatory diseases such as sepsis. In a breakthrough study of 2015, SHAO's group identified a novel GSDMD protein as a common substrate for activated capase-1 and caspase-4/5/11 in innate immunity. Cleavage of GSDMD by caspase induces pyroptosis (Shi et al., Nature 2015). Following this work, WANG-DING's group collaborated with SHAO's group, and proved that GSDMD is the pyroptosis executioner. Activated caspases cleave the linking loop between the N- and C-terminal domain of GSDMD, thereby unlocking the autoinhibition on the GSDMD-N domain which then translocates to plasma membrane through binding to membrane lipids and perforates the membrane to cause pyroptosis (Ding et al., Nature 2016). Despite these progress, the mechanism for how caspases in innate immunity, especially caspase-4/5/11, are activated by upstream signals, and how activated caspases specifically target GSDMD to trigger pyroptosis is unknown. This is an emerging important question in pyroptosis studies.
Both caspase-1 and caspase-4/11 belong to a large conserved family of cysteine proteases, named caspase. The caspase family has attracted decades of hot studies due to their close association with apoptosis. All caspases are synthesized as inactive zymogens containing a conserved protease domain divided into a large p20 and a small p10 subunit; their activation requires cleavage between the p20 and the p10 which generate a p20/p10 heterodimer. In this study, researchers discovered that LPS-stimulated caspase-4 also undergoes autoprocessing in vitro and in treated cells. Unlike caspase-11 which autoprocesses after Asp285 at the N-terminus of p10, caspase-4 autocleaves at two sites, Asp270 at the C-terminus of p20 and Asp289 (corresponding to Asp285 in caspase-11) at the N-terminus of p10. Mutagenesis analyses further confirmed that Asp289/285 in caspase-4/11 is the only autoprocessing site critical for their activation by cytosolic LPS for cleaving GSDMD and inducing pyroptosis.
The dogma in caspase biochemistry is that all caspases recognize a tetrapeptide motif xxxD (D for aspartate, x for any residue) within a substrate and cleave after the aspartate. Extensive peptide substrate profiling has established the tetrapeptide sequence preferred by each caspase. Caspases in apoptosis often cleave a dozen of substrates. However, caspase-4/11 only have one known physiological substrate GSDMD, and caspase-1 cleave two more cytokines IL-1β/18 besides GSDMD. The caspase dogma cannot explain why caspases in innate immunity bear such a narrow substrate spectrum. Surprisingly, researchers found that the GSDMD-C domain, but not the cleavage-site tetrapeptide, mediates the recognition of GSDMD by activated caspase-4/11. They successfully determined the crystal structures of the complexes between autoprocessed caspase-4/11 and GSDMD-C domain. The complex structures show that autoprocessed caspase-4/11 p20/p10 further dimerize to form a heterotetramer. At the dimerization interface, a two-strand intermolecular β-sheet distant from the catalytic site cysteine, extends outwards and shapes like a key inserted into a preformed hydrophobic groove in the GSDMD-C domain. Each caspase-4/11 heterotetramer contains two symmetric intermolecular β-sheets which mediate binding of two GSDMD-C molecules, resulting in a 2:2 enzyme-substrate complex. The caspase-4/11-GSDMD complex structures also demonstrate that N-terminal extension of p10 would introduce inappropriate structural clashes to GSDMD-C domain, explaining why caspase-4/11 need to be precisely processed at Asp289/285. Mutagenesis analyses confirmed the hydrophobic interface is the structural basis for caspase-GSDMD recognition and GSDMD-cleavage-mediated pyroptosis. Researchers further investigated the caspase-1 recognition of GSDMD. Crystal structure of caspase-1-GSDMD-C complex shows a similar GSDMD-recognition mode as the case of caspase-4/11.
This study provides complete and thorough understanding of caspase-1/4/11 autoprocessing and recognition of pyroptosis executioner GSDMD in innate immunity. The reported caspase-GSDMD complex structures are the first structures of enzyme-substrate complex in caspase studies. The findings of this study provide in-depth insights into substrate recognition and enzymatic mechanism of caspase family. Caspases have important functions in cancer and immunity and are potential drug targets. However, it has been challenging to obtain lead compounds that are sufficiently selective for a particular caspase given structural conservation in the catalytic pocket among all caspases. Thus, the GSDMD-binding hydrophobic pocket in caspase-1/4/11, which is outside of the catalytic site, suggests a new space for developing caspase drugs to treat pyroptosis-caused inflammatory diseases such as sepsis.
The research work was supported by the Strategic Priority Research Program of CAS, Basic Science Center Project and Excellent Young Scholar Program of NSFC, the National Key Research and Development Programs of China, the program Initiative for Innovative Medicine of Chinese Academy of Medical Sciences, and the program Youth Innovation Promotion Association of CAS.
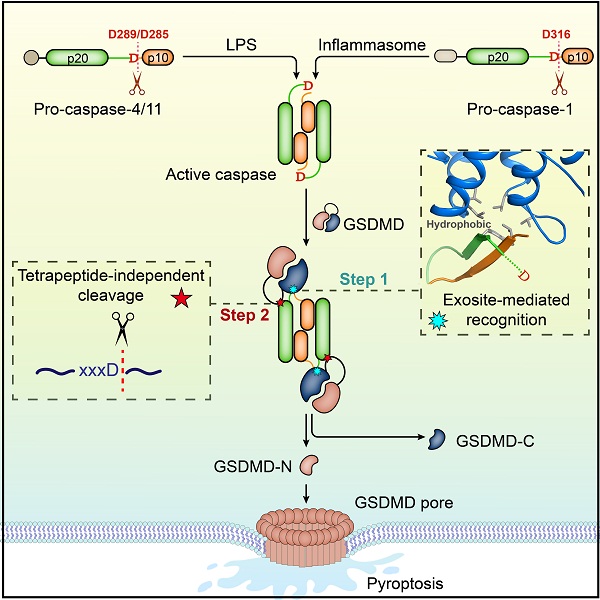
Schematic diagram of the molecular mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis.
(Image by Dr. DING Jingjin's lab)
The web link for this paper is https://www.sciencedirect.com/science/article/pii/S009286742030146X
Contact: Jingjin Ding, Ph.D.
Institute of biophysics, Chinese Academy of Sciences
Beijing100101, China
Tel: (86)-10-64888548
Email: jding@ibp.ac.cn
(Reported by Dr. DING Jingjin's group)

