Structural basis for the biosynthesis of long-chain alk(a/e)nes in photosynthetic cyanobacteria
The demands for petrol-based fuels has remained high, hence biofuels are of particular interest as a renewable supplement for gradually diminishing fossil fuels. Long chain alk(a/e)nes are the major constituents of conventional transportation fuels, hence biosynthesis of alkanes represents a promising way to produce substitutes for petroleum-based fuels.
An earlier work discovereda pathway responsible for alkane biosynthesis in cyanobacteria, which allows the conversion from solar energy to alkanes through photosynthesis, without requiring of external carbon sources. This two-step pathway utilizes two water-soluble enzymes, namely acyl-acyl carrier protein (acyl-ACP) reductase (AAR) and aldehyde deformylating oxygenase (ADO) (Fig.a). AAR catalyzes the first step of this pathway, reducing the substrate fatty acyl-ACP/fatty acyl-coenzyme A (acyl-CoA) into corresponding aldehyde in a NADPH-dependent manner. Subsequently, ADO catalyzes the conversion of fatty aldehyde into alk(a/e)ne with one carbon shorter. The catalytic and regulative mechanisms of this pathway have not been elucidated yet. Moreover, due to the poor solubility of long chain aldehydes, it has been remained mysterious how are the aldehydes transferred between two water-soluble enzymes in the hydrophilic environment.
On Mar 23, 2020, a research article entitled "Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase" was published in Nature communications by Professor LI Mei's group from the Institute of Biophysics, Chinese Academy of Sciences. They reported the crystal structure of AAR from Synechococcus elongatus PCC7942 in its apo form, together with three structures of AAR in complex with ADO, in which AAR binds either substrate or substrate/cofactor (Figs. b & c).
The structural and biochemical results revealed details about the binding mode of AAR with ligands, provided structural basis for the "PING PANG" mechanism adopted by AAR. The AAR-ADO complex structure showed the interaction pattern between the two enzymes, allowed the identification of key residues for the complex formation, and unraveled a long contiguous channelrunning through both enzymes, which could be the potential aldehyde transferring pathway from AAR to ADO.
This work provides structural basis for the alkane biosynthesis pathway in cyanobacteria (Fig. d), laying a solid foundation for further optimizing the pathway to increase the alkane yield, and engineering cyanobacteria for the biofuel production.
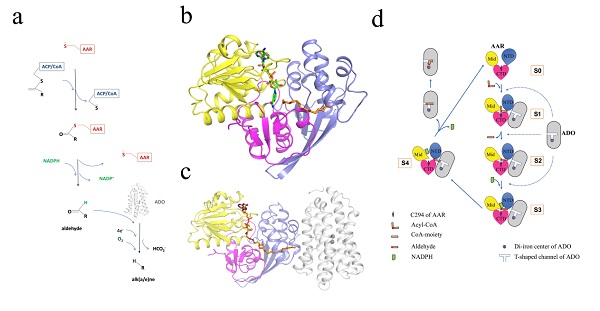
Figure legend: Enzyme structures and catalytic process of cyanobacterial alk(a/e)ne biosynthesis pathway. (a)Scheme of alk(a/e)ne biosynthesis pathway catalyzedby AAR and ADO. (b) Structure of AAR. Three domains of AAR are shown in different colors. (c)Structure of AAR-ADO complex. ADO is shown in white. (d) Cartoon representation of the proposed catalytic process of the alkane biosynthesis pathway in cyanobacteria based on the present study.
(The image by Dr. LI Mei's lab)
The web link for this paper is https://www.nature.com/articles/s41467-020-15268-y
Contact: LI Mei
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Phone: 86-10-6488511
Email: meili@ibp.ac.cn
(Reported by Dr. LI Mei's group)

