Trimming longer RNA transcripts to become micro RNA – key mechanism revealed
In living organisms, genetic information stored in DNA, or genes, is first transcribed into messenger RNA by RNA polymerase II, and then the RNA transcripts direct the synthesis of proteins, which play the vast majority of the tasks in life processes. However, not all RNA transcripts are destined for making proteins, and those that do not code for protein synthesis are called non-coding RNAs (ncRNAs). A particular group of ncRNAs, known as microRNAs (miRNAs), are ~22 nucleotide (nt) in length and they play vital roles in diverse physiological processes, including regulation of gene expression, cell differentiation and development. Aberrant expression of miRNAs has been linked to the development and progression of various human diseases including cancers. Certain miRNAs are even considered as biomarkers for diagnosis and targets for drug discovery, making it of great significance to study the mechanisms of miRNA biogenesis and regulation.
The RNA transcripts destined for generating miRNAs have a characteristic stem-loop structure of ~65 nt in length, and the first step of becoming a functional miRNA is to have the extra RNA bits beyond the stem-loop region removed by a protein complex called Microprocessor in the cell nucleus. This first RNA cleavage reaction generates a precursor miRNA (pre-miRNA) that still retain the RNA loop, and the pre-miRNA is then exported to the cytoplasm where it undergoes a second cleavage reaction to produce mature miRNA. Despite great many progresses in the past decade, detailed mechanisms concerning the first cleavage reaction, including how pri-miRNA is recognized by Microprocessor, and how Microprocessor cuts at the correct sites on pri-miRNA, remain poorly understood. In the work entitled "Structural Basis for pri-miRNA Recognition by Drosha", published online in the journal Molecular Cell on March 27, 2020, a team of researchers led by Dr. XU Rui-Ming from Institute of Biophysics, Chinese of Academy of Sciences, and Dr. WANG Hong-Wei from Tsinghua University has determined a near atomic-resolution structure of Microprocessor bound to pri-miRNA, and the work provided adequate answers to the mechanistic questions outlined above.
The Microprocessor complex is composed of Drosha, the catalytic subunit for RNA cleavage, and DGCR8, an RNA binding protein important for pri-miRNA binding. The researchers reconstituted the Microprocessor complex using purified proteins produced in insect cells, and pri-miRNA by in vitro transcription. The reconstituted RNA-protein complex or the protein complex alone were studied by single-particle cryo-electron microscopy to reconstruct the three-dimensional structure of the biomacromolecular specimen.
They found that Drosha plays an especially important role in pri-miRNA recognition and specification of the cleavage sites, in agreement with results from previous studied. And they identified three key domains of Drosha that cooperatively recognize pri-miRNA, the PAZ domain, MB helices and the double-stranded RNA binding domain (dsRBD). The PAZ and MB helices together recognize the single- to double-stranded junction region of pri-miRNA, and this interaction is critical for Drosha to bind pri-miRNA in the correct orientation and cut pri-miRNA at correct sites. Meanwhile, the dsRBD binds the upper stem region of pri-miRNA, stably positioning the RNA stem against to the catalytic center of the enzyme for cleavage reaction. They also discovered that the PAZ domain of Drosha binds both strands of RNA at the internal region, instead of the 3' termini bound by all other PAZ domains known to date. Furthermore, they observed a large conformational change of the PAZ domain before and after RNA binding, suggesting an auto-regulated mechanism of RNA binding of Drosha, which may be beneficial for distinguishing specific targets from an abundance of various forms of RNA inside the cell nucleus.
In sum, this work solved a long-standing problem of how Drosha recognizes a pri-miRNA and binds it in a correct mode to allow precise cleavage of pri-miRNA. The results provide fundamental understandings to the mechanisms of miRNA biogenesis. The study was supported by grants from the National Natural Science Foundation of China, Ministry of Science and Technology of China, Beijing Municipal Science and Technology Commission, and Chinese Academy of Sciences.
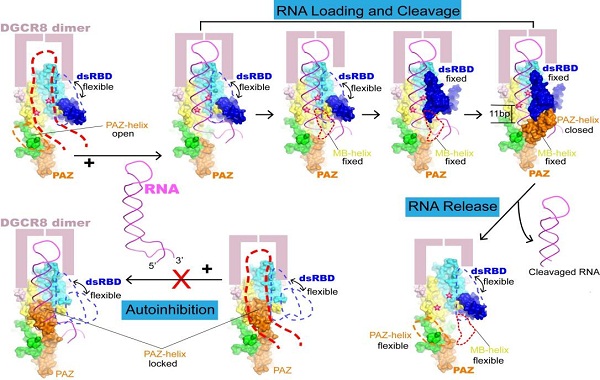
A model for how Drosha/DGCR8 complex recognizes pri-miRNA substrates and specifies the cleavage sites
(Image by Dr. XU Ruiming's group)
Article link: http://www.sciencedirect.com/science/article/pii/S1097276520301441
Contact: XU Ruiming
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: rmxu@ibp.ac.cn
(Reported by Dr. XU Ruiming's group)

