Research team led by CAS Researcher Wang Xiaoqun co-creates cell map and gene regulatory network in developing human hippocampus
Decoding the development of the human hippocampus was published online by Nature on January 16, 2020. This work identifies regulatory networks and key markers of specific gene expression in the developing human hippocampus, provides a high-precision illustration of relevant cells, and analyzes cell types, key markers and transcriptional regulatory networks in the development process.
The hippocampus is an important part of the limbic system in the human brain that has essential roles in information coding, spatial navigation and the consolidation of information from short-term memory to long-term memory. In addition, given its high relevance to pathogenesis of multiple diseases such as epilepsy, mental disorders and Alzheimer's disease, the human hippocampus is an area of interest to clinicians and neuroscientists. Research has found that one of the first areas in the brain affected by Alzheimer's disease is the hippocampus, with early symptoms including impairment of memory and disorientation. Development of the human hippocampus is essential to our research on understanding cellular and molecular mechanisms in memory formation. Though the hippocampus is an evolutionarily conserved part of vertebrates, existing understanding of cellular and molecular features in the developing human hippocampus is insufficient.
For the purpose of this research, the team has used single-cell RNA sequencing and assay for transposase-accessible chromatin using sequencing (ATAC–seq) analysis to illustrate the cell types, cell linage, molecular features and transcriptional regulation of the developing human hippocampus. Using the transcriptomes of 30,416 cells from the human hippocampus at gestational weeks 16–27 with help of tSNE, they have identified 47 cell subtypes in 8 types and their developmental trajectories. In addition, key neurons development was studied systematically, and the cellular and molecular mechanism in memory formation circuit and the development process of the human hippocampus were further clarified. The research also compared molecular features of the human hippocampus and those of the mouse and revealed gene expression differences between the two species. This work comprehensively reveals the timeline and key genes in hippocampus development. Overall, it provides a blueprint for understanding human hippocampal development and is a tool for investigating related diseases.
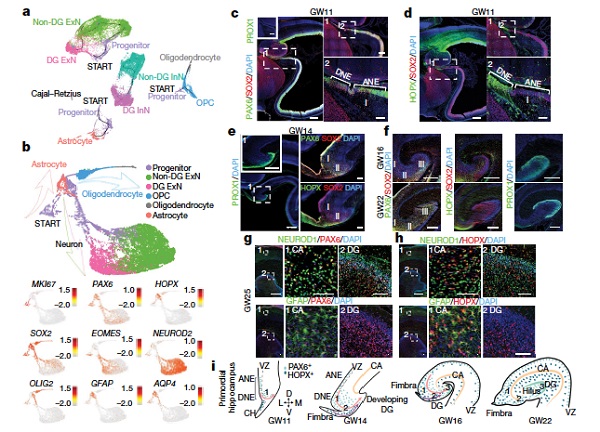
Fig. Human hippocampus development paths and key regulatory molecules
This work is a collaboration between the Institute of Biophysics of Chinese Academy of Sciences, Beijing Normal University, and Peking University. CAS Institute of Biophysics Researcher Wang Xiaoqun and Prof. Wu Qian of Beijing Normal University are co-correspondents. Co-first authors include Dr. Zhong Huijuan, Associate Researcher Sun Le, Doctoral Candidate Lu Yufeng of CAS Institute of Biophysics and Dr. Ding Wenyu of Beijing Normal University. It was supported by the National Basic Research Program of China by Ministry of Science and Technology ("973 Program"), the CAS Strategic Priority Research Program, and the National Natural Science Foundation of China, among others.

