The Mechanism of New Spacer Acquisition in CRISPR-Cas System Discovered by IBP Scientists
Bacteria and Archaea regularly face the threat of invading foreign DNA, such as phages, plasmids, transposons and so on. To protect themselves from the invading genetic elements, prokaryotes have evolved many strategies. One of the most widespread is the CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR associated) systems.
The CRISPR-Cas system processes the adaptive immunity through three steps. PhaseⅠcalled Spacer acquisition, short foreign-derived sequence which termed “spacers” are selected and incorporated into a CRISPR array from the invading element by the core proteins Cas1 and Cas2. In phaseⅡ (expression), spacers are transcribed into a pre-crRNA and then processed into mature crRNA which binds Cas proteins and forms the ribonucleoprotein complex. In the last interference phase, the crRNA guides the resulting surveillance complex to recognize and degrade the invading DNAs complementary to crRNA. In recent years, the molecular mechanisms of the expression and interference phase were well-characterized, however the spacer acquisition is the least understood aspect.
Memory of the invading nucleotide acid is the key feature of the CRISPR-Cas system. The re-invasion of the foreign element will trigger the memory bank to defend the invaders with the “acquired information”. Therefore, studying on acquisition phase is of great significance to understand the whole CRISPR-Cas system.
In a deep study, WANG Jiuyu, LI Jiazhi and colleagues (led by Prof. WANG Yanli at the Institute of Biophysics of Chinese Academy of Sciences) solved the crystal structures of Cas1-Cas2 with a series of DNAs, revealing the spacer acquisition step is PAM-dependent and provide significant structural basis of the adaptive immunity mechanism.
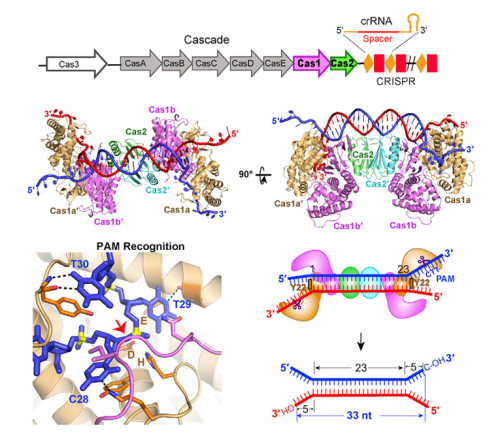
The crystal structures of Cas1-Cas2-DNA complexes revealed that two Cas1 dimers sandwich a Cas2 dimer forming a flat DNA binding surface. Only the DNA in dual-forked configuration can be selected to be the protospacer, and the 3’-overhangs of the prostospacer are essential for the new spacer acquistion. This study also highlighted that each Cas1 dimer has one catalytic subunit, which recognize the PAM sequence to select the correct protospacer, and cleave the 3’-overhangs to generate two 3’-OH groups.On the base of discovering the exact DNA substrate used by Cas1-Cas2 in vivo, the team illuminated that the architecture of the Cas1-Cas2 predetermined the length of newly acquired spacer, and the Cas1-Cas2complex undergoes large conformational change upon the DNA bidinging, just like a butterfly dropping its wings from “wings-up” to “wings-down”. Building on the detailed messages provided by the structures of the various Cas1-Cas2-DNA complexes, Cas1 and Cas2 show their specific roles in spacer acquisition and how they recognize the PAM complementary sequence via sequence-specific interactions, which shed light on the molecular basis for the selection of the protospacer with PAM. Together, this structural research provides critical insights into the molecular mechanism of the adaptation process, which will significantly facilitate the research on this adaptive immunity system and pave the way towards better development and utilizing of the CRISPR-Cas system.
The work entitled “Structural and Mechanisitic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems” was published online by the Cell on Oct. 15, 2015.
The research was funded by Natural Science Foundation of China (91440201), the Chinese Ministry of Science and Technology (2014CB910102), and the Strategic Priority Research program of the Chinese Academy of Sciences (XDB08010203). The X-ray diffraction data were collected at beamlines BL-17U, BL-18U, and BL-19U at the Shanghai Synchrotron Radiation Facility.
Figure: Crystal Structure of E. coli Cas1-Cas2 Bound to Dual-Forked protospacer DNA. Cas1-Cas2 recognizes and binds to a Dual-Forked DNA containing PAM-Complementary sequence, then predetermined the protospacer length prior to inserting it into CRISPR-Cas locus of E. coli. ( ? cell)
Contect
WANG Yanli
Institute of Biophysics, Chinese Academy of Sciences
Website: http://english.ibp.cas.cn/ibp_pi/W/201312/t20131213_114402.html
Email: ylwang@ibp.ac.cn