FUS interacts with ATP synthase beta subunit and induces mitochondrial unfolded protein response
A research article lead by Prof. WU Ying at IBP, entitled “FUS interacts with ATP synthase beta subunit and induces mitochondrial unfolded protein response in cellular and animal models” was published online by PNAS on September 24, 2018. This study provides the first evidence that a predominantly nuclear RNA binding protrin (RBP) targets mitochondrial ATP synthase. It has significant implications for future development of diagnostic tools and therapeutic approaches to age-related neurodegenerative diseases.
FUS (fused in sarcoma/translocated in liposarcoma,FUS/TLS), a DNA/RNA binding protein, is predominantly localized in nucleus, but is also known to shuttle between the nucleus and the cytoplasm. FUS protein has been associated with multiple steps in RNA metabolism, including RNA transcription, RNA splicing and microRNA processing. FUS-proteinopathies, a group of heterogeneous neurodegenerative disorders including Amyotrophic Lateral Sclerosis with FUS pathology (ALS-FUS) and Frontotemporal Lobar Degeneration with FUS pathology (FTLD-FUS), are characterized by the formation of inclusion bodies containing the nuclear protein FUS in the affected patients. So far, more than 50 mutations in the FUS gene have been found in patients with FUS-proteinopathy. However, the underlying molecular and cellular mechanisms of FUS in these diseases remain unclear.
Previous work from WU lab has established a drosophila model of FUS-proteinopathy, which could recapitulate major clinical and pathological features of these devastating diseases (Chen et al., 2011 Protein Cell). Based on the animal model along with cellular models and FTLD-FUS patients’brain samples, they found that FUS interacted with mitochondrial chaperon HSP60 and HSP60 mediated FUS localization to the mitochondria, leading to mitochondrial damage and finally neuronal cell death. Mitochondrial impairment may represent a potential therapeutic target for treating FUS proteinopathies (Deng et al., 2015 PLoS Genetics).
In this new study, the authors further demonstrated that mitochondrial dysfunction occurs as the earliest detectable change induced by FUS. In cellular and fly models, FUS interacts with the mitochondrial ATP synthase-subunit (ATP5B), disrupts ATP synthase complex assembly, suppresses the activity of mitochondrial ATP synthase, and activates the mitochondrial unfolded protein response (UPRmt). ATP5B expression is increased in cells and flies expressing FUS. Down-regulating expression of ATP5B or UPRmt genes ameliorates FUS-induced neurodegeneration. These data uncover a previously unknown role of FUS in targeting mitochondrial ATP synthesis and activating UPRmt.
Drs. WU Ying and ZHU Li are the corresponding authors. Drs. DENG Jianwen and WANG Peng from Dr. WU’s lab are the co-first authors of this paper. This work was funded by NSFC grants.
Article link:
http://www.pnas.org/content/early/2018/09/20/1806655115
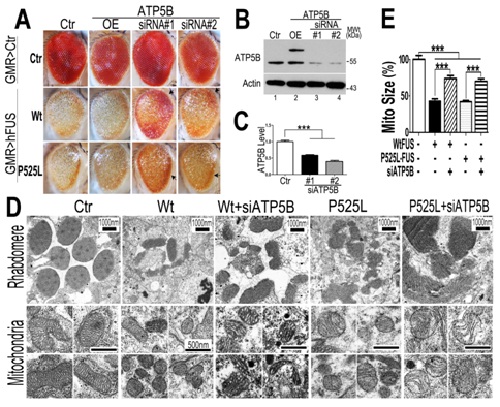
Figure 1. Down-regulating ATP5B ameliorates retinal degeneration and mitochondrial fragmentation in flies expressing Wt or P525L-mutant FUS.

Figure 2. Working model for FUS-induced mitochondrial damage and neurotoxicity.
Contact: ZHU Li
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Phone: 86-10-64888303
Email: zhuli@ibp.ac.cn

