WANG Yanli group and ZHANG Xinzheng group reveal the molecular mechanism of Type III CRISPR-Cas system
CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats, CRISPR-associated genes) are RNA-guided adaptive immune systems that protect most archaea and approximately half of bacteria against invading foreign nucleic acids. Type III CRISPR-Cas systems are characterized by the presence of Cas10 protein and are subdivided into four (III A-D) subtypes. Types III-A system comprises a Csm effector complex, which is composed of five different Csm proteins (Csm1-5) and one crRNA. The crRNA-guided complexes recognize complementary RNA targets and cleave them via the Csm3 subunit. The Csm1, also called Cas10, possesses palm domain and HD domain, which is responsible for cOA synthesis and DNA cleavage respectively.
In the paper titled "Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference" published in Cell on November 29, 2018, two groups (Prof. WANG Yanli and Prof. ZHANG XinZheng) from Institute of Biophysics, Chinese Academy of Sciences, reported the crystal structure of Csm, and a series of cryo-EM structures of Csm in complex either with cognate or non-cognate RNA targets. These structures revealed that the mechanisms of crRNA-guided RNA cleavage, and the target RNA binding dependent DNA cleavage and cOA synthesis.
You et al. showed that the Csm complex is composed of five Csm subunits (Csm1-5) and one crRNA with a protein stoichiometry of Csm1122334151. The spacer region of the crRNA form base pairs with the complementary target RNA, forming crRNA-target RNA duplex with every 6th base being flipped out. This study showed Csm3 proteins cleave the target RNA periodically. Importantly, the comparison of cognate RNA or non-cognate RNA bound Csm complexes showed that the 3' anti-tag region of target RNA has two distinct binding channels depending on complementarity between the 5'-tag of crRNA and 3' anti-tag of target RNA. Additionally, Csm1 subunit undergoes a conformational change upon the cognate target RNA binding, allosterically activating the DNA cleavage and cOA synthesis.
These structural studies provide crucial insights into the mechanistic processes required for both crRNA-meditated, sequence-specific RNA cleavage, as well as transcription-dependent, non-specific DNA cleavage and cOAs generation, which will significantly facilitate the research on type III CRISPR-Cas system.
Graduated student You Lilan, Associate Prof. Ma Jun and Wang Jiuyu are co-first authors in this paper. All the cryo electron microscopy work was performed in Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences. The X-ray diffraction data were collected at the BL-17U1, and BL-19U1 beamlines at SSRF. This work was supported by grants from the Chinese Ministry of Science and Technology, the Natural Science Foundation of China, the Strategic Priority Research Program of the Chinese Academy of Sciences and HHMI-Wellcome.
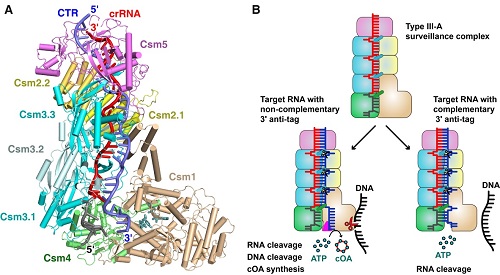
Structure Studies of the CRISPR-Csm Complex.
(A) Overall structure of Csm-cognate RNA -AMPPNP complex.
(B) Model of Type III-A CRISPR-Cas interference.
Article link: https://www.cell.com/cell/fulltext/S0092-8674(18)31451-X#%20
Contact: Yanli Wang
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101,China
Phone: 86-10-64881316
Email: ylwang@ibp.ac.cn
(Reported by Dr. Wang Yanli’s group)

