WANG Yanli group reveals the molecular mechanism of CRISPR–SpyCas9 inhibition by a phage anti-CRISPR protein
Almost all archaea and half of bacteria possess Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated genes (Cas) adaptive immune systems, which protect microbes from the foreign nucleic acids. The prokaryotic CRISPR-Cas systems are divided into two classes. Class 1 systems possess multi-subunit effector complexes comprised of multiple Cas proteins, whereas Class 2 systems are characterized by effector complexes that consist of a single, large Cas protein. The Cas9, Cas12a, Cas12b and Cas13 proteins of Class 2 systems have been successfully harnessed for genome-editing tools.
CRISPR-Cas systems provide bacterial immunity against invading phages by crRNA-guided nuclease activity. To evade bacterial defenses, phages express Acr proteins that inhibit CRISPR-dependent responses. Two AcrIIA proteins AcrIIA2 and AcrIIA4 from a Listeria monocytogenes phage were found to inhibit the activity of Streptococcus pyogenes Cas9 (SpyCas9) in bacterial and human cells. Molecular mechanism of Cas9 inhibition by AcrIIA4 has been well studied. While both AcrIIA2 and AcrIIA4 were found to inhibit Cas9 by disrupting target double-stranded DNA (dsDNA) binding, the precise molecular mechanisms by which these two anti-CRISPR proteins block target DNA binding are different. Recent structural studies revealed that AcrIIA4 disables Cas9 by DNA mimicry. However, how AcrIIA2 inhibits Cas9 remains to be elucidated.
In the study published online in Molecular Cell on Dec. 31, 2018, entitled Phage AcrIIA2 DNA Mimicry: Structural Basis of the CRISPR and Anti-CRISPR Arms Race, Dr. LIU Liang and colleagues (led by Prof. WANG Yanli at the institute of Biophysics of Chinese Academy of Sciences) solved the crystal structure of the AcrIIA2-Cas9-sgRNA ternary complex at 3.3 angstrom resolution. AcrIIA2 is positioned in a positively charged cleft within SpyCas9 that is formed by the PI, HNH, WED, and REC2 domains. Structural comparison between the AcrIIA2-bound and target-dsDNA-bound SpyCas9-sgRNA complexes shows that AcrIIA2 binds SpyCas9 at a position similar to the target DNA binding region. More specifically, AcrIIA2 interacts with the protospacer adjacent motif (PAM) recognition residues of Cas9, preventing target double-stranded DNA (dsDNA) detection. Thus, phage-encoded AcrIIA2 appears to act as a DNA mimic that blocks subsequent dsDNA binding by virtue of its highly acidic residues, disabling bacterial Cas9 by competing with target dsDNA binding with a binding motif distinct from AcrIIA4. This study provides a more detailed mechanistic understanding of AcrIIA2-mediated inhibition of SpyCas9, the most widely used genome-editing tool, opening new avenues for improved regulatory precision during genome editing.
This research was supported by grants from the Natural Science Foundation of China, the Chinese Ministry of Science and Technology, and the Strategic Priority Research Program of the Chinese Academy of Sciences. The X-ray diffraction data were collected at the BL-17U1, BL-18U, and BL-19U1 beamlines at the National Center for Protein Sciences Shanghai (NCPSS) at SSRF and the BL41XU beamline at SPring-8.
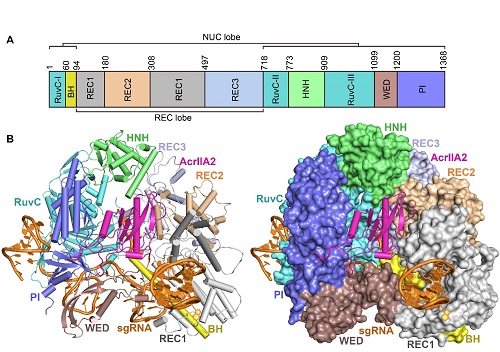
The crystal structure of the AcrIIA2-Cas9-sgRNA ternary complex
(A) Domain organization of SpyCas9.
(B) Ribbon and surface representations of the AcrIIA2-SpyCas9-sgRNA complex.
Article link: https://www.cell.com/molecular-cell/fulltext/S1097-2765(18)30981-X
Contact: Yanli Wang
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101,China
Phone: 86-10-64881316
Email: ylwang@ibp.ac.cn
(Reported by Dr. Wang Yanli’s group)

