IBP Scientists Determined the Structure of C2c2, Effector of Type VI CRISPR-Cas System
Almost all archaea and half of bacteria possess Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated genes (Cas) adaptive immune systems, which protect microbes from the foreign nucleic acids. The prokaryotic CRISPR-Cas systems are divided into two classes. Class 1 systems possess multi-subunit effector complexes comprised of multiple Cas proteins, whereas Class 2 systems are characterized by effector complexes that consist of a single, large Cas protein. The Cas9 and Cpf1 proteins of Class 2 systems have been successfully harnessed for genome-editing tools.
C2c2 is a newly identified effector of Class 2 and type VI CRISPR-Cas system. In contrast to the Cas9, Cpf1 and C2c1 proteins, C2c2 does not have any known DNA nuclease domain, but has two conserved HEPN domains, which are commonly associated with ribonucleases. C2c2 then was found to be a single-component programmable RNA-guided RNA-targeting CRISPR effector, protecting the host from the RNA viruses. In addition, C2c2 is able to cleave the pre-crRNA, generating the mature crRNA. Given its dual RNase activities, C2c2 may be used to develop new RNA-targeting tools. Therefore, the molecular mechanism of pre-crRNA processing as well as crRNA-guide single stranded RNA cleavage by C2c2, is important to develop C2c2 based RNA tools.
In the study published online in Cell on Jan. 12, 2017, entitled Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities, Dr. LIU Liang and colleagues (led by Prof. WANG Yanli at the institute of Biophysics of Chinese Academy of Sciences) solved the crystal structure of Leptotrichia shahii C2c2 in its crRNA-free and crRNA-bound states. LshC2c2 exhibits a bilobed structure, which differs fundamentally from Cas9 and Cpf1, both in architecture and domain organization. The two lobes of Lshc2c2 are rearranged upon the crRNA loading to form a central channel. LIU Liang et al. found that the two catalytic pockets responsible for cleaving pre-crRNA and target RNA are located on the Helical-1 and HEPN domains, respectively. They also found that the HEPN-catalytic site on the outer surface is located distantly from the crRNA guide region, accounting for promiscuous target cleavage.
These structural studies provide critical insights into the molecular mechanism of the dual RNase activities of C2c2, which will significantly facilitate the research on this type VI CRISPR-Cas system and pave the way towards better development and utilizing of the RNA editing tool.
This research was supported by grants from the Natural Science Foundation of China, the Chinese Ministry of Science and Technology, and the Strategic Priority Research Program of the Chinese Academy of Sciences. The X-ray diffraction data were collected at the BL-17U1, BL-18U, and BL-19U1 beamlines at the National Center for Protein Sciences Shanghai (NCPSS) at SSRF and the BL41XU beamline at SPring-8.
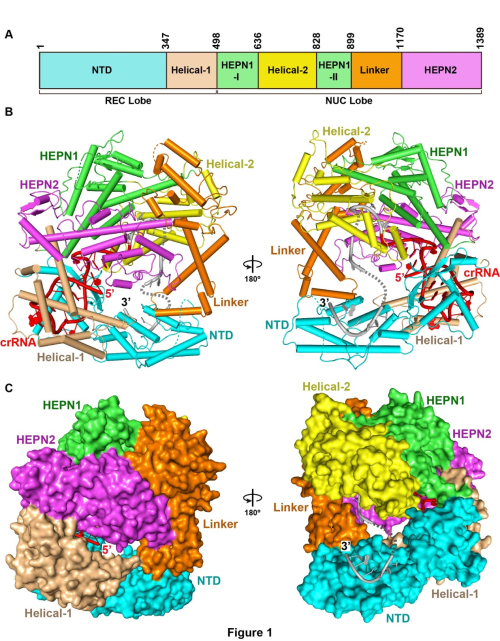
Figure 1. The Crystal Structure of LshC2c2-crRNA Binary Complex (Image by IBP)
Contact:
WANG Yanli
Institute of Biophysics
Email:ylwang@ibp.ac.cn
Tel: 86-10-64881316
Fax:86-10-64881316

