Professor Fuquan Yang’s group conduct the first large-scale phosphoproteome analysis of rat L6 myotubes
Post-translational modifications play a key role in the regulation of intracellular biological processes, such as the cell cycle, apoptosis, metabolism, cellular signaling and proliferation. Protein phosphorylation is one of the most widespread and important protein post-translational modifications, with about 30% of all proteins in eukaryotic cells being phosphorylated at any given time.Identifying phosphoproteins and their phosphorylation sites is important for understanding the role that protein phosphorylation plays in a particular cellular process. Mining as much information as possible from phosphoproteomes, of both a static and a dynamic nature, is thus a key task which will facilitate greater understanding of biological events that are regulated by phosphorylation.
Professor Fuquan Yang’s group is developing new methods and techniques for high-throughput, sensitive identification of post-translational modifications, such as protein phosphorylation, and also for membrane proteomics, quantitative proteomics and lipidomics. In a recent paper in the Journal of Proteome Research1 they used rat L6 myotubes as an in vitro model system to study the role of phosphorylation in signaling pathways in skeletal muscle. They conducted global phosphoproteome profiling of rat L6 myotubes using an efficient mass spectrometry-based phosphoproteomic strategy which included prefractionation of tryptic peptide mixtures with self-packed RP C18 columns, phosphopeptide enrichment with TiO2, and 2D-LC (SCX/RP)-MS/MS analysis. A total of 2230 unique phosphopeptides from 1195 proteins were identified using a target/decoy database searching strategy, with a false-discovery rate of less than 1.0%. 11 Ser motifs and one Thr motif were identified within the data set using the Motif-X algorithm, and several potential signaling pathways, such as the MAPK signaling pathway and the IGF-1/Insulin signaling pathway, were found in this myotubes phosphoproteome.
This primary qualitative phosphoproteome of resting myotubes should provide a basis and resource for further studies on the role of protein phosphorylation in the molecular mechanisms that regulate cell signal transduction.
1Hou et al. (2010) Phosphoproteome analysis of rat L6 myotubes using reversed-phase C18 prefractionation and titanium dioxide enrichment. J. Proteome Res. 9: 777–788
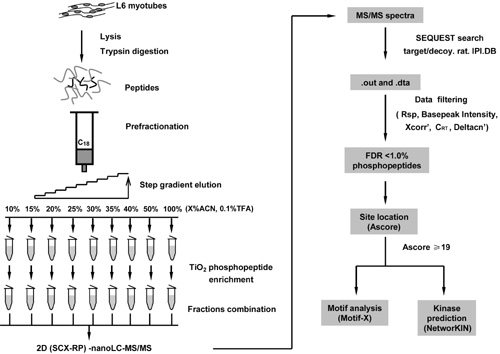
Fig. Strategy used for the large-scale phosphorylation analysis of rat L6 myotubes.

