Sun Fei's team reveals key molecular mechanism in S-OPA1-mediated mitochondrion intimal fusion
On April 14, 2020, Sun Fei's team from the National Laboratory of Biomacromolecules, CAS, published its latest research on eLife: Cryo-EM structures of S-OPA1 reveal its interactions with membrane and changes upon nucleotide binding. This work focuses on how S-OPA1 (soluble short isoform in the Dynamin family) is combined and assembled in mitochondrial inner membrane, and identifies the structure and assembly of S-OPA1 induced by nucleotide binding.
Mitochondria, which are double-membrane organelles, can form remarkable dynamic networks through membrane fusion and fission regulated by dynamins. However, the double-membrane structure poses challenges in understanding the molecule mechanism in its fusion and division. OPA1 is a key protein in mediated mitochondrion intimal fusion. Under physiological conditions, OPA1 undergoes proteolytic processing to form a membrane-anchored long isoform (L-OPA1) and a soluble short isoform (S-OPA1). A combination of L-OPA1 and S-OPA1 is essential for efficient membrane fusion. When cells are under certain pressure, L-OPA1 can be cleaved into S-OPA1, which suppresses the mitochondrial inner membrane infusion. Recently, also from the National Laboratory of Biomacromolecules, CAS, Hu Junjie’s team published structural analysis of a trimeric assembly of the mitochondrial dynamin-like GTPase Mgm1 on PNAS (https://www.pnas.org/content/117/8/4061), which elaborates how S-Mgm1 mediates mitochondrial inner membrane infusion and cristae shaping. However, the molecular mechanism by which S-OPA1 mediates mitochondrial inner membrane infusion remains unclear.
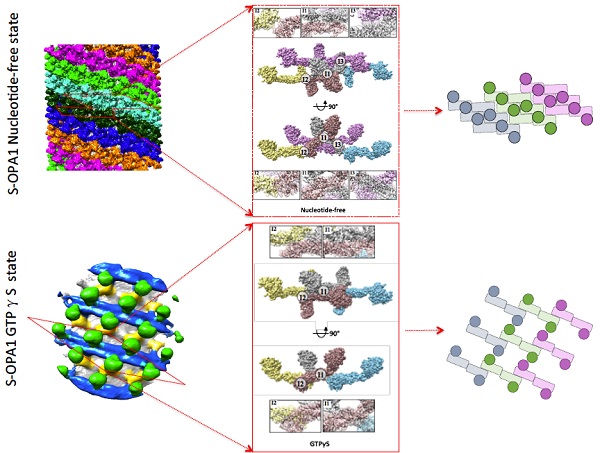
The team used a three-dimensional reconstruction technique of cryo-electron microscopy (cyro-EM) for helical reconstruction formed by cleaved S-OPA1 and liposome. S-OPA1 has a classic dynamin-like structure that contains a G/BSE domain, a stalk domain, and an EMB domain. The helical reconstruction processing procedure indicated that a S-OPA1–coated tube contains six helical starts. It is also observed that the asymmetric unit of the S-OPA1 packing array contains two S-OPA1 molecules. There are four interaction surfaces between adjacent asymmetric units, i.e., I1, I2, I3 and P1. Such compact packing suggests that stalk interactions play a pivotal role in maintaining the structural stability of the S-OPA1-lipid complex. A mutagenesis experiment verifies that the S-OPA1 EMB domain is responsible for membrane binding and deformation. Further mutagenesis experiments support that S-OPA1 can induce tubulation of liposomes without the addition of nucleotide, suggesting that the tubulation activity of S-OPA1 is independent of its GTP hydrolysis activity. Relevant results can confirm that S-OPA1 can deform membranes without requiring GTP hydrolysis and further suggest that GTP hydrolysis of S-OPA1 most likely occurs after liposomal tabulation.
Cryo-ET and SVA were used to analyze the assembly of GTPγS-bound S-OPA1 on the membrane. Compared to the nucleotide-free state, after GTPγS binding, considerable gaps appeared between stalks in neighboring helical rungs as their interval expanded, which led to reduced compactness of the S-OPA1 assembly and a loosened helical lattice. Diameter of tubes increased, contrary to reducing tube diameter caused by Dynamin family protein as observed previously. This is highly relevant to the OPA1-induced membrane fusion.
Sun Fei, researcher at the National Laboratory of Biomacromolecules, CAS, is the correspondence author. From his team, Zhang Danyang (PhD) and Zhang Yan (associate research fellow) are co-first authors.
The team thanks Prof. Edward H Egelman from the University of Virginia for his generous help on image processing of helical reconstruction. All the EM works were performed at the Center for Biological Imaging, Institute of Biophysics, CAS. Special thanks go to the Center for support on EM sample preparation and data collection.
Article Link: https://doi.org/10.7554/eLife.50294
(Reported by Dr. SUN Fei's group)

