Xue Yuanchao's research group develops a new RIC-seq technology to analyze RNA in situ advanced structures and targets
A research paper entitled "RIC-seq for global in situ profiling of RNA–RNA spatial interactions" was published in Nature on May 6, 2020. In the paper, a new RIC-seq (RNA in situ conformation sequencing) technology that discovers RNA in situ advanced structures and intermolecular RNA–RNA interactions in cells was developed, the conformation and organization of mRNA and noncoding RNA in HeLa cells was analyzed, the whole genome enhancer-promoter RNA regulatory network map was drawn, and a new mechanism for how enhancer RNA activates oncogene MYC transcription was clarified.
RNA structure regulates the transmission of genetic information. For example: the dynamic changes of the snRNA structure in the spliceosome ensure efficient and precise splicing; the ribosomal RNA advanced structures are necessary for translation; the pseudoknot structure of the telomerase RNA component (TERC) ensures telomerase activity and chromosome stability. In cells, most RNAs form secondary structures through intramolecular base pairing, and fold into complex tertiary structures under the guidance of RNA binding proteins. The highly structured RNA molecules then interact with each other for biological regulation, so it is very important to analyze RNA in situ advanced structures and targets in cells. Besides, the existing high-throughput sequencing technologies for RNA secondary structures and intermolecular interactions cannot effectively discover RNA advanced structures, and the use of in vitro proximity ligation leads to an overly high false positive rate.
In response to the above issues, a new RIC-seq technology was used to generate in situ three-dimensional (3D) interaction maps of RNA in HeLa cells, revealing the spatial organization of protein-encoding mRNA and noncoding RNA. As to its application, this paper found that the interaction between enhancer and promoter-associated noncoding RNAs can be used to infer its regulatory network, and analyzed the new mechanism for how the binding of super-enhancer long noncoding RNA CCAT1-5L, RNA-binding protein, MYC promoter and enhancer RNA changes chromatin conformation, thereby regulating oncogene MYC transcription.
According to the paper, the RIC-seq technology can not only detect RNA advanced structures, but also accurately identify the targets of various types of noncoding RNAs, which facilitates in-depth study of the "structural code" and functionality of non-coding RNA. In addition, the technology can also be used for the study of viral RNA structure and targets.
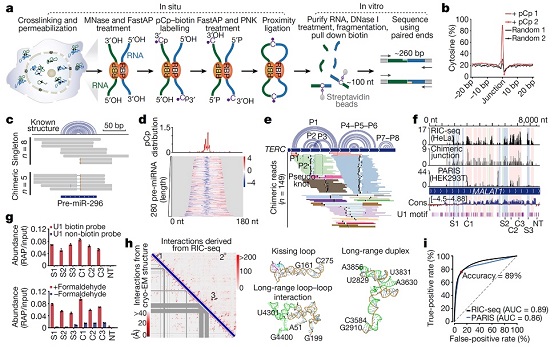
Figure. RIC-seq can accurately discover RNA in situ advanced structures and targets.
The study was completed by the Institute of Biophysics, Chinese Academy of Sciences, Henan Normal University and Xinyang Normal University. Ph.D. Student Cai Zhaokui, Associate Research Fellow Cao Changchang and Ji Lei of the Institute of Biophysics, Chinese Academy of Sciences are co-first authors of this paper, and Research Fellow Xue Yuanchao is the corresponding author. The study was supported by the National Natural Science Foundation of China Noncoding RNA Research Program, National Key R&D Program, Strategic Pilot Research Program of the Chinese Academy of Sciences, etc.
Link to article: https://doi.org/10.1038/s41586-020-2249-1
(Reported by Dr. Xue Yuanchao's group)

