Gao Pu and Zhang Xinzheng Groups Jointly Reveal the Assembly, Catalysis and Regulatory Mechanisms of Type I R-M System
The paper "Structural insights into assembly, operation, and invasion of a Type I restriction modification system" by the Gao Pu and Zhang Xinzheng groups was published online by Nature Microbiology on June 1, 2020. This work researches the classic Type I R-M system with EcoR124I.
A variety of complex samples of the EcoR124I system (R2M2S1/R1M2S1/M2S1) with targeted DNA and two phage encoded proteins (Ocr and ArdA) were successfully prepared. The freeze electron microscopic structures of 10 different physiological conformations of the system were analyzed, and the mechanism of assembly, catalysis and regulation of the system was revealed by a large number of biochemical experiments (Fig. 1).
Fig. 1: Host Cells’ Immune Recognition of and Response to Pathogenic DNA
In brief, this work has the following highlights: (1) it is the first time that the Type I R-M system has precisely controlled different enzyme activities by forming different conformational states, which were completely unknown before; (2) it reveals that Type I R-M system can accurately control different enzyme activities by forming different conformational states. Most of the intermolecular interaction interfaces found in this work are reported for the first time; (3) the phage proteins Ocr and ArdA inhibit Type I R-M in the way of DNA mimics. However, these two inhibitors do not affect the conformational changes of the system; (4) this work is not limited to solving a structural biology problem that has been puzzling for decades; more importantly, it reveals many previously unreported mechanisms of action.
In conclusion, this study systematically reveals the molecular mechanism of assembly, catalysis and regulation of the Type I R-M system, which advances understanding of the role of this system in resisting phage infection and other physiological functions of bacteria, and provides a basis and ideas for designing biological tools and phage therapy based on the Type I R-M system.
The Restriction Modification (R-M) system, known as the innate immune system of bacteria, can methylate the host's own DNA and cut the unmodified foreign DNA. As the earliest identified and most sophisticated R-M system, the Type I R-M system has a variety of subunit assembly modes, and has DNA methyltransferase, DNA translocase and DNA endonuclease activities. Identification of the Type I R-M system promoted the discovery of other types of R-M systems and triggered the revolution of molecular genetics. The research and application of restrictive enzymes also won the Nobel Prize in physiology and medicine in 1978. Despite more than half a century of extensive research, little is known about how the Type I R-M system is assembled, how it achieves multi enzyme activity conversion, and how it is regulated by phage proteins. In addition, due to the complex and frequent conformational changes of the system itself, obtaining high-resolution structural information of various states has always been a challenging task in the field.
Immunological recognition of the Pathogenic DNA is an important means for host cells to perceive pathogenic infection. This immune response exists widely from lower prokaryotes to higher mammals (Fig. 1). Mammalian cells recognize pathogenic DNA and mediate immune responses through multiple innate immune pathways; bacteria mainly rely on the "R-M system" and "CRISPR CAS system" to remove invasive phage DNA.
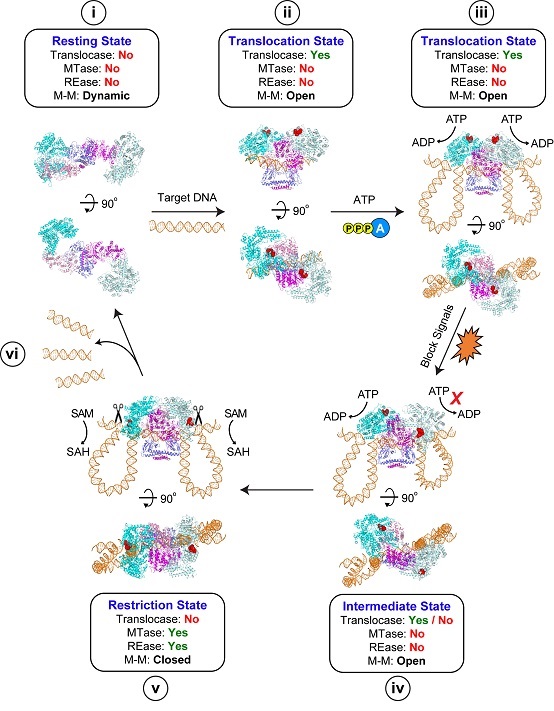
Fig. 2: Working Model of Type 1 R-M holoenzyme complex
The web link for this paper is https://www.nature.com/articles/s41564-020-0731-z
(Reported by Dr. GAO Pu's group)

