CAS researchers unveil molecular mechanism of autophagy formation
A paper titled "Multi-site-mediated entwining of the linear WIR-motif around WIPI β-propellers for autophagy" was released on the online-only journal Nature Communications on June 1, 2020.
The paper, submitted by a team led by Feng Wei, a researcher at the Institute of Biophysics, Chinese Academy of Sciences, unveils the molecular mechanism through which WIPI proteins recognize and bind to ATG2 and regulate autophagy formation with the help of biochemistry and structural biology.
Autophagy is a highly-conserved self degradation pathway in eukaryotic cells. It plays an important role in maintaining homeostasis, avoiding accumulation of harmful proteins and resisting invasion of pathogens through degradation and recycling of cellular contents. The WIPI protein family is the effector molecule of PI3P lipid, which can recruit other factors to regulate autophagy formation. The WIPI family proteins can specifically recognize a linear sequence (WIR-Motif) of ATG2 protein to form a WIPI /ATG2 complex, located at the junction of phagocytic vesicles and endoplasmic reticulum, and promotes autophagy extension and closure. However, the recognition patterns between them and the mechanism of autophagy formation are unclear.
On the basis of previous studies on WIPI protein (JMB, 2019), Feng's team obtained a stable WIPI3 / ATG2A-WIR-Motif protein complex by fusion expression, and analyzed its high-resolution crystal structure. The structure showed that the ATG2A-WIR-Motif protein "twined" itself at the bottom and the rim of the disc-like WIPI3 protein. Their interaction involves multiple sites on the WIPI protein. However, amino acid mutation at the binding site will destroy the formation of WIPI / ATG2 complex, and affect the occurrence of autophagy. It is noteworthy that many known disease-related point mutations are just distributed on the interaction interface, and relevant biochemical experiments also show that the introduction of these point mutations will destroy the interaction between WIPI protein and ATG2, which explains the pathogenic mechanism of these point mutations. This study also proposed a novel substrate recognition pattern mediated by "multiple sites" of the WIPI protein, and defined the amino acid sequence characteristics of WIPI3 / 4 protein recognition, providing an important basis for further study of the WIPI protein's role in regulating autophagy.
With Feng as the corresponding author, the paper has associate researcher Ren Jinqi, Dr Liang Ruobing and assistant researcher Wang Wenjuan as its co-first authors. Dr Zhang Dachuan and Professor Yu Li of Tsinghua University also contributed greatly to the paper. The research was jointly funded by the Key R & D Program of the Ministry of Science and Technology, the Class-B CAS Pilot Project and the National Natural Science Foundation of China.
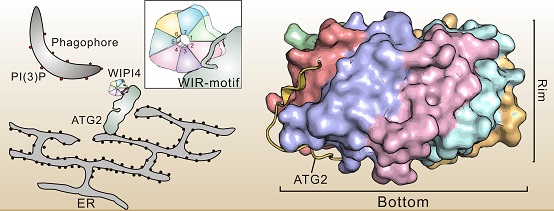
Molecular mechanism of WIPI protein recognition and binding to ATG2
The left sketch shows that WIPI recognizes and binds to a linear sequence (WIR-Motif) of ATG2 protein to form a WIPI /ATG2 complex located at the junction of phagocytic vesicles and endoplasmic reticulum, promoting the extension and closure of autophagy. The high-resolution crystal structure of the WIPI3/ATG2A-WIR-Motif complex is shown on the right.
The web link for this paper is https://doi.org/10.1038/s41467-020-16523-y
(Reported by Dr. FENG Wei's group)

