Chinese scientists achieve breakthrough in protein synthesis
The research described in the paper was done by the teams of Zhou Xiaolong and Wang Enduo from the CAS Center for Excellence in Molecular Cell Science and that of Chen Chang from CAS’s Institute of Biophysics.
The human genome encodes 38 aminoacyl tRNA synthetases (aminoacyl-tRNA synthetase, aaRS), which are responsible for two protein synthesis systems, cytoplasmic and mitochondrial. The aaRS catalyzes the aminoacylation of tRNA to ensure the speed of protein synthesis while the catalytic editing reaction ensure its accuracy. The speed and accuracy are regulated by nutrition, environment and pathogen infection. For example, when bacterial infection occurs, human cytosolic methionine tRNA synthetase (MetRS) is phosphorylated, and some non-tRNAMet are mistakenly aminoacylated, so that methionine is mistakenly referred to in the non-methionine codon to mediate adaptive translation; under oxidative stress, cysteine (Cys) in the active site of bacterial threonide synthetase (ThrRS) can be modified by sulfosulfonate, resulting in loss of editing and mistranslation. Salmonella enterica phenylalanyl-tRNA synthetase (PheRS) can produce oxidative modification under oxidative stress, which promotes an editing reaction. However, the potential regulatory effect of nitrification stress on protein synthesis has long been undiscovered.
The research found that oxidative stress induced by reactive oxygen species H2O2 cannot regulate the activity of ThrRS in mitochondria and cytoplasm of humans or mice; on the contrary, GSNO, a nitric oxide donor of reactive nitrogen specifications (RNS), significantly downregulates the aminoacylation and misaminoacylation of mitochondrial ThrRS. With the irreversible biotin-switch procedures(IBP) combined with mass spectrometry (MS), four Cys residues were identified with S-nitrosation modification. Among them, Cys409 is the amino acid activation center. It was found that the nitrosylation of each Cys residue down regulated the activity of aminoacylation and editing to varying degrees through the construction of a series of mutants, combined with biochemical analysis. Furthermore, eight kinds of mitochondrial aaRS and 14 kinds of cytoplasmic aaRS were identified from human mitochondria and mouse cortex tissues respectively. Further study found that the mitochondrial ThrRS nitrosylation in cortical tissue was significantly reduced in the Huntington disease mouse model, suggesting that the regulation of mitochondrial protein synthesis may play a potential role in diseases.
The study discovered the inhibitory effect of nitrification stress on ThrRS aminoacylation and editing reaction, drew the aaRS nitrosylation modification network of cytoplasm and mitochondria in human cells and mouse cortical tissues for the first time, and revealed the regulation of nitrosylation modification on protein synthesis speed and accuracy in mammalian cells, providing new clues regarding the molecular mechanism of Huntington neurodegenerative diseases.
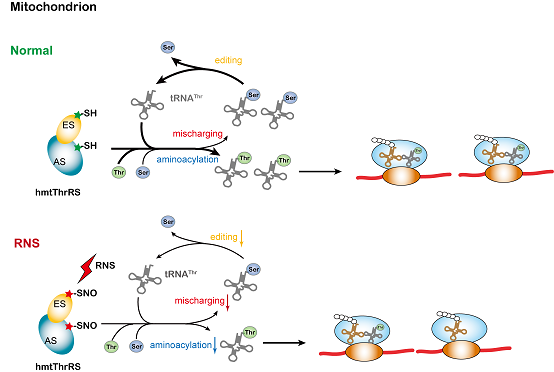
The figure shows the regulatory effect of RNS-mediated nitrosation on mitochondrial ThrRS activity.
The web link for this paper is https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkaa471/5850316
(Reported by Dr. CHEN Chang's group)

