Dr. ZHU Bing's group dissected the kinetics and mechanisms of mitotic DNA methylation maintenance and their contributionsin aging-associated methylome deterioration
On June 24, 2020, Cell Research published a research article entitled "Kinetics and mechanisms of mitotic inheritance of DNA methylation and their roles in aging-associated methylome deterioration"online. In this work, the authors dissected the dynamic process and underlying mechanisms of DNA methylation maintenance, and revealed the contribution ofdifferentialmaintenance efficiency inselective methylation loss during aging and tumorigenesis.
DNA methylation is an epigenetic modification involved in regulating the spatiotemporal gene expression programs, X chromosome inactivation, Genomic imprinting and repetitive element repression. In mammalian cells, DNA methylation mainly occurs at the C5 position of CpG dinucleotides, and the methylation pattern should bemaintainedin each round of mitotic cell division, which is critical for cell identity maintenance. However, due to technology limitation, the kinetics, underlying mechanisms and fidelity of methylation maintenance are not well studied.
In this work, Bing Zhu's group developed a method named Hammer-seq to study the kinetics of DNA methylation maintenance at single-molecular level. Hammer-seq combined EdU labeling, biotin conjugation, immunoprecipitation and hairpin bisulfite sequencing technologies. With the advantage of simultaneously detecting methylation status of the same CpG sites in the parent and daughter strands, Hammer-seq can not only measure methylation maintenance kinetics, but also measure the potential accompanying de novo methylation events.
They found methylation maintenance is indeed highly efficient, as maintenance ratio is greater than 50% in minutes after replication fork passage, which supports the existence of a replication-coupled maintenance phase. On the other hand, methylation maintenance continues hours after fork passage, which indicates the presence of a replication-uncoupled maintenance phase.
By combining Hammer-seq with systematically mutating various factors, they found DNMT1-PCNA and UHRF1-LIG1 interactions contribute to the replication-coupled rapid methylation maintenance, UHRF1-H3K9me2/3 interaction and nucleosome occupancy regulate the replication-uncoupled methylation maintenance, and LSH preferentially facilitates replication-uncoupled methylation maintenance in late S-replicating regions. Meanwhile, replication-uncoupled phase is sufficiently robust enough to largely restore the methylome when replication-coupled maintenance is disturbed, suggesting the potent maintenance capacity reserved in replication-uncoupled phase. For different genomic contexts, high methylated CpG density regions show both higher maintenance rate and de novo methylation frequency.
Aged tissue samples and cancer cells that experiencing extensive mitotic divisions show hypo-methylation at global level and hyper-methylation at selective CpG sites. However, the underlying mechanisms remain largely unclear. In this work, we found CpG sites (such as solo-WCGW) displaying methylation loss in aging and tumorigenesis exhibit both low maintenance rate and low de novo methylation frequency, suggesting low maintenance efficiency is an potentially important contributor to methylation loss in aging and tumorigenesis. Thus, although mitotic inheritance of DNA methylation is highly robust, it remains to be imperfect, and CpG sites with low maintenance efficiency will inevitably loss methylation after extensive mitotic divisions in aging and tumorigenesis.
Prof. ZHU Bing is the corresponding author. Dr. MING Xuan and Dr. ZHANG Zhuqiang are the co-first authors. ZOU Zhuoning, Dr. DONG Qiang, HE Qixiang, YI Yangyang and Dr. LI Yingfeng from ZHU Bing laboratory made important contributions in Cell line establishment and/or data analysis. LV Cong performed the UHPLC-MS/MS experiments under the supervision of Prof. WANG Hailin. This work is supported by the National Natural Science Foundation of China, the Ministry of Science and Technology of China, and the Chinese Academy of Sciences.

Fig1:Flowchart of Hammer-seq, a novel approach to measure DNA methylation maintenance within single DNA molecule level.
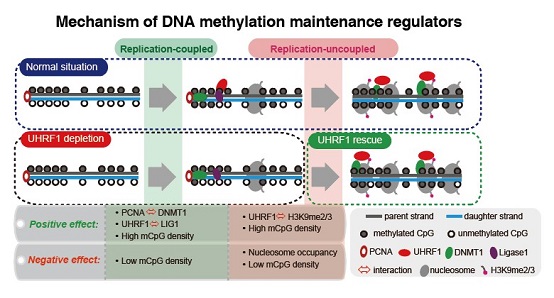
Fig2:Mechanisms of DNA methylation maintenance regulators.
(Image by Dr. ZHU Bing’s group)
The web link of this paper: https://www.nature.com/articles/s41422-020-0359-9
Contact: ZHU Bing
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: zhubing@ibp.ac.cn
(Reported by Dr. ZHU Bing’s group)

