Research team reveals the novel catalytic regulatory mechanism of ubiquitination and deubiquitination mediated by pathogens
On June 2, 2020, the journal Nature Communications published online the research paper "Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors" by Gao Pu's team. This work analyzes the complex structure of a novel ubiquitinating enzyme MavC derived from the highly pathogenic Legionella pneumophila and the substrate protein UBE2N and ubiquitin molecules, and simultaneously analyzes the two complex structures of MavC and its homologous protein MvcA and negative regulator Lpg2149. Combined with a large number of biochemical and cellular experiments, it explains the molecular mechanism of the catalysis and regulation of this new ubiquitination modification system.
Protein ubiquitination modification is one of the most important post-translational modifications in eukaryotic cells. The classic ubiquitination modification needs to be catalyzed by the E1-E2-E3 three-enzyme cascade reaction, while the two recently identified species of L. pneumophila effector proteins (SidE and MavC) can rely on catalyzing the ubiquitination modification of host proteins in a completely new way independent of E1-E2-E3 (Figure 1) (Figure 1). Gao Pu's team once reported the structure and molecular mechanism of the catalytic reaction of SidE family proteins in “Cell” magazine in 2018, and this latest work mainly focuses on the research of MavC family proteins.
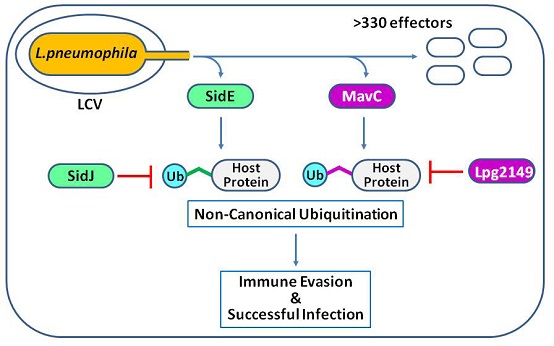
Figure 1: Two novel ubiquitination modification systems encoded by pathogens
The genome cluster where MavC is located can encode three effector proteins: Lpg2147 (MavC), Lpg2148 (MavC paralog A or MvcA) and Lpg2149. Both MavC and MvcA can catalyze the deamination of ubiquitin molecules (Ub-Q40 to Ub-E40), while Lpg2149 can inhibit the two’s activity. Recent studies have shown that MavC can also specifically ubiquitinate the host protein UBE2N, thereby inhibiting the normal E2 function of UBE2N and the activation of the NF-κB pathway. Interestingly, although MvcA is very similar to MavC, MvcA cannot catalyze UBE2N ubiquitination.
The researchers used MavC to catalyze the preparation of ubiquitinated UBE2N (UBE2N~Ub), and analyzed the crystal structure of the complex between MavC and UBE2N~Ub (Figure 2a). The structure directly reveals the interaction interface between MavC and UBE2N and Ub, and shows that MavC uses the same active center to catalyze the deamination reaction and the ubiquitination reaction. The sequence and structure comparison of MvcA and MavC shows that there is no conservative presence of key amino acids that can bind UBE2N in the Insertion domain of MvcA. That also explains why MvcA cannot stably bind UBE2N in the free state and why it cannot catalyze the ubiquitination modification of UBE2N. These conclusions based on the structure have been further verified through biochemical and cell experiments.
Unexpectedly, MavC and MvcA also have deubiquitinating enzyme activity, which can specifically remove MavC-mediated UBE2N ubiquitination modification (Figure 2b). Since MvcA has stronger deubiquitination activity than MavC, MavC and MvcA are likely to play the roles of ubiquitinase and deubiquitinase in actual infection, respectively. This finding indicates that MavC can use the same active pocket to catalyze the three enzymatic reactions of deamination, ubiquitination and deubiquitination. At the same time, MavC is identified as the first enzyme to be able to simultaneously add and remove the same type of ubiquitination modification.
The researchers further analyze the crystal structures of the MavC+Lpg2149 complex and the MvcA+Lpg2149 complex (Figure 2a). Unlike the crystal structure in the free state showing a tight dimer, Lpg2149 interacts with MavC or MvcA as a monomer. Since Lpg2149 and Ub occupy similar binding positions, it indicates that Lpg2149 can directly interfere with the interaction between Ub and MavC/MvcA. Functional experiments show that Lpg2149 can indeed simultaneously inhibit deamination, ubiquitination and deubiquitination. Interestingly, the free state of MavC itself has the binding conformation of Lpg2149, while MvcA requires a large conformational change to achieve the binding with Lpg2149. Further binding experiments show that the affinity of MavC-Lpg2149 is about 50 times stronger than that of MvcA-Lpg2149, which is consistent with the structural analysis.
In summary, this study clarifies the catalysis and regulation mechanism of Mavc-mediated new ubiquitination modification, which not only helps to understand the diversity mechanism of the game between pathogen and host, but also provides ideas for the design of new anti-bacterial therapies.
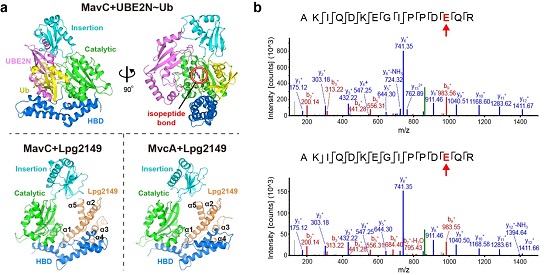
Figure 2: a, crystal structures of the three complexes of MavC+UBE2N~Ub, MavC+Lpg2149 and MvcA+Lpg2149; b, cleavage sites of the MavC (top) and MvcA (bottom) catalyzed UBE2N~Ub deubiquitination

