Analyzing chromatin status of poised genes may provide a powerful tool to predict the transcription potential
In eukaryotic cells, the genomic DNA is hierarchically packaged by histones into chromatin. Chromatin structure and its plasticity play an important role in the activation and silencing of gene transcription, and determine the cell fate during differentiation and development. Therefore, it is an important issue for scientific frontier to study the chromatin status and transcription potentials of the key development genes during the cell differentiation.
Based on epigenetic markers and chromatin binding proteins, the chromatin states are divided into many different types, such as active, repressive, and poised (or bivalent) states. The poised state is defined by simultaneous presence of two opposing chromatin modifications, active trimethylation of histone3 lysine 4 (H3K4me3) and repressive trimethylation at histone 3 lysine 27 (H3K27me3). The poised state correlates strongly with pluripotency in ESCs, with the potential of rapid gene activation at the differentiation programs. However, unlike epigenetic modifications, the structural characteristic of poised chromatin remains largely unknown, nor does its correlation with gene expression potential.
A study published online in Cell Reports with the title of "Analysis of Local Chromatin States Reveals Gene Transcription Potential during Mouse Neural Progenitor Cell Differentiation", revealed the structural characteristics of poised genes, and provided a powerful tool to predict the genes' transcription potential.
Employing a mild time-course MNase-seq approach (TC-rMNase-seq), the researchers identified two distinct groups of MNase-sensitive chromatin status in mouse embryo stem (mES) cells, i.e. "active" (Region I) and "poised" (Region II) chromatin regions, which show opposing MNase-seq trends during the digestion time course. On this basis, they developed a computational method to quantify chromatin status and gene expression potential. Using a parameter COPI (Chromatin Opening Potential Index), they successfully scored the transcriptional potentials of related genes in mES cells according to the different accessibilities at their promoters. To explore the utility of the method, they validated that the genes with high COPIs, such as Lfng, Ypel1, Scrt2 and Ildr2, are poised in mES cells, however their promoters are partially opened and are readily activated during neuron progenitor cells (NPCs) differentiation. Among them, Lfng and Scrt2 are known to function in NPCs and morphogenesis respectively. More importantly, through COPI prediction, the researchers identified two novel factors, Ildr2 and Ypel1 also play important roles during the mouse NPC differentiation in vitro. They proposed that COPI scores provide a powerful tool to predict and identify important novel genes involved in a variety of biological processes, including key regulators during development and differentiation, and intermediate early genes in neural and immune responses.
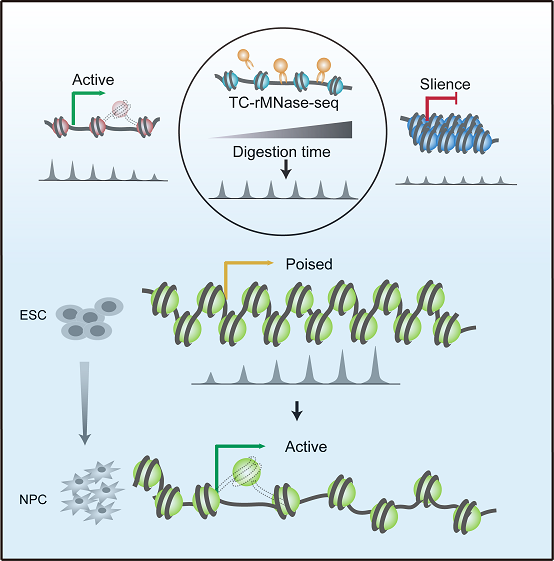
Figure. Model for the prediction of gene expression potential through identifying the local chromatin states
(Image by Dr. LI Guohong’s group)
This research was finished by a research team led by Prof. LI Guohong from Institute of Biophysics (IBP) of the Chinese Academy of Sciences (CAS) and Prof. XU Xueqing from Baoan Maternal and Child Health Hospital of Jinan University.
Article link: https://www.sciencedirect.com/science/article/pii/S2211124720309347
Contact: LI Guohong
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: liguohongt@ibp.ac.cn
(Reported by Dr. LI Guohong’s group)

