Single-Cell Transcriptome Analysis Reveals Cell Lineage Specification in Temporal-Spatial Patterns in Human Cortical Development
As the core component of the human central nervous system (CNS), the cerebral cortex has human-specific advancement in evolution. Understanding the temporal and spatial characteristics of cerebral cortex development at the single-cell level help us to understand the molecular regulatory mechanism of the formation of complex cortical structures and functions.
On Aug, 21th, 2020, Professor WANG Xiaoqun of the institute of Biophysics (IBP) in cooperation with Professor TANG Fuchou and Professor QIAO Jie of the Peking University,published the research article entitled "Single-cell transcriptome analysis reveals cell lineage specification in temporal-spatial patterns in human cortical development" in the journal Science Advances.
The researchers systematically analyzed the neuronal developmental process in the four representative cortical lobes including the frontal lobe (FL), parietal lobe (PL), occipital lobe (OL), and temporal lobe (TL) and the subcortical region pons from gestational weeks 9 to 26 using RNA sequencing. They identified 8 major cell type and their corresponding subclusters at the developmental stage and performed functional verification for critical cell types. Benefit from rigorous study design and systematic sampling, the researchers found a novel neural progenitor subpopulation that was transiently abundant in the SVZ region at GW11-14. Further analysis of specific gene expressions and signal pathways in this subpopulation showed that, these cells highly expressed neural stem cell and neuronal genes, as well as components of canonical WNT signaling pathway. They maintained proliferation activity and supposed to further generate deep-layer neurons.
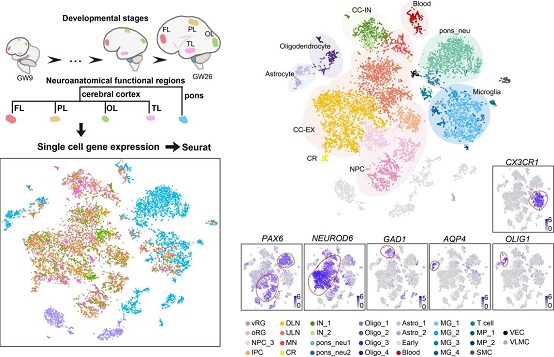
Spatiotemporal single-cell analysis of gene expression in human developing cerebral cortex together with pons.
(Image by Dr. WANG Xiaoqun's group)
Then, the researchers analyzed the differentially expressed genes and developmental trajectory within these neural progenitor subtypes. In this dataset, they performed functional verification for the long intergenic noncoding RNA (lincRNA) which is specifically expressed by the outer radial glia (oRG). oRG cells present in large numbers at outer subventricular zone (oSVZ) of human and non-human primate development but rare in SVZ of mouse development, which is thought to underlie massive production of cortical neurons and the evolution from lissencephalic to gyrencephalic brain. According to scRNA-seq analysis, the researchers observed that the lincRNA LINC00943 was specifically expressed by oRG cells. Overexpressing LINC00943 in the fetal mice cerebral cortex significantly increased oRG cells in the SVZ region, indicating that this lincRNA played an important role in maintenance of neural subpopulation and regional specification.
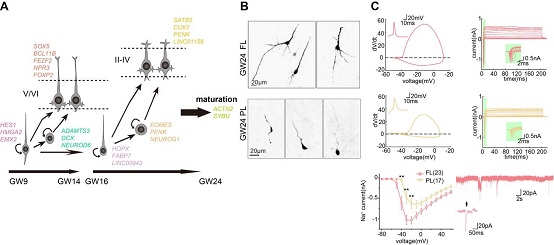
The developmental trajectory of neural progenitor and functional verification of LINC00943 in oRG population maintenance during mouse cortical development
(Image by Dr. WANG Xiaoqun's group)
In this dataset, the researchers summarized temporal sequences of heterogeneous neuronal appearance and molecular characteristics of neuronal development. They first detected the neuron alaction potentials (APs) in prefrontal lobes at GW24, indicating that neurons in FL exhibited earlier electrophysiological properties and more complex morphology that other cortical lobes. These evidences revealed the temporal differences of neuronal development in different cortical regions by electrophysiological recording. In addition, regional comparisons of the whole transcriptome revealed neurons from the cerebral cortex and the subcortical region, the pons, showing marked distinctions in both molecular regulations and developmental pattern. By comparing the cerebral cortex to the pons, they defined cellular and molecular features of cortical neurons.
This study provided a comprehensive resource for neuronal development of human embryonic and fetal cerebral cortex, including pairwise regional information, which would be important for decoding the highly complex yet highly robust developmental trajectory of the human brain. Prof. WANG Xiaoqun, Prof. TANG Fuchou and Prof. QIAO Jie are co-corresponding authors. Co-first authors include Dr. FAN Xiaoying, FU Yuanyuan, Dr. ZHOU Xin and Dr. SUN Le. This work was supported by the National Natural Science Foundation of China (NSFC), the National Basic Research Program of China, Chinese Institute for Brain Research, Beijing and Beijing Advanced Innovation Center for Genomics at Peking University.
The web link of this paper: https://advances.sciencemag.org/content/6/34/eaaz2978
Contact: WANG Xiaoqun
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: xiaoqunwang@ibp.ac.cn
(Reported by Dr. WANG Xiaoqun's group)

