IBP Team Reports Application of Active RNA Interference in Mitochondria
The mitochondria team at the Key Laboratory of RNA Biology of the Institute of Biophysics (IBP), the China Academy of Sciences (CAS), published an article entitled "Active RNA interference in mitochondria" on Cell Research on August 17, 2020. The article details the IBP team's discovery of RNA interference in mitochondria (mitoRNAi) and its application in mitochondrial research.
In recent years, thanks to advances in cell biology and molecular biology, as well as a deeper understanding of mitochondria functions, it is known that many diseases induced by mitochondrial function defects, such as deafness, Leber hereditary optic neuropathy (LHON), NARP syndrome, Leigh syndrome, MELAS syndrome, MERRF syndrome, and cardiomyopathies, are caused by mutation in mitochondrial genes. To address the treatment needs on relevant diseases, mtDNA research is a key focus of fundamental sciences, medicine development and clinical treatment. It is a pressing issue for us to figure out how to effectively edit mtDNA and regulate the genome expressions.
ZFN, TELEN and CRISPR systems are the mainstream technologies currently in use for genome editing. Due to difficulty in transporting sgRNA into mitochondria through its double membrane, the widely-used CRISPR system is not an ideal tool when editing genome in mitochondria. On the other hand, mitoTALENs and mtZENs, both developed to address the sgRNA transporting, are dependent on repeated protein domains for identifying base sequences during mtDNA editing. Recently, Prof. David R. Liu of Harvard University proved that using TLENS, the base sequences of the same type can be interchanged within certain domains. Additionally, as in every single cell there may be hundreds to tens of thousands of mitochondria in constant fusion and fission, this kind of editing is subject to losses and hard to sustain homogeny effect.
In 2014, the team of Dr. ZHANG Xiaorong discovered evidence for the presence of miRNA and its effector protein Argonaute2 (Ago2) within the mitochondria, and their role in enhancing mitochondrial genome expression during myogenesis. Their research shows that miRNA can enter the mitochondria, and that Ago2 indeed resides within the mitochondria, as tested by Trypsin protection analysis (Zhang et al., 2014, Cell). Their findings unveiled a new mechanism of miRNA regulation, and a new layer to the regulation on mitochondrial genome expression.
RNAi effect was first discovered in mitochondria at the beginning of this century. Its presence was later identified in yeast, trypanosome, drosophila melanogaster, plants and mammals. As of now, its mechanism of action is clearly understood, and there are extensive applications in this regard, mostly on cytoplasm and nucleus. However, it remains unclear if RNAi exists in semiautonomous organelles such as mitochondria. As siRNA and miRNA are of a similar size and act similarly through Ago2 protein mediation, siRNA may also play a regulating role in mitochondria.
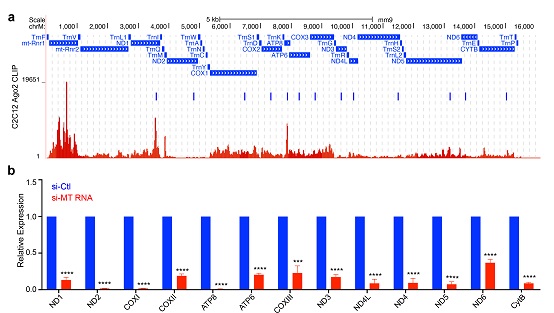
Fig 1: Applying mitoRNAi in mtDNA-encoded transcripts. (a). siRNA designed to target individual mtDNA-encoded transcripts according to previously mapped Ago2-binding peaks in C2C12 cells. (b) RT-QPCR analysis of all 13 mitochondrial transcripts in C2C12 cells after treatment with corresponding siRNs.
(Image by IBP's Mitochondrial Biology Team)
Based on these, we developed a strategy called "mitoRNAi" that involves knocking down targeted gene transcript of mitochondria. Using this tool, we validated that siRNA is capable of entering the mitochondria via the RNA enzyme protection test, immunofluorescence co-localization test, and ClickIn test. But more needs to be done to understand how siRNA enters the mitochondria. Based on Ago2-clip data, we improved the siRNA design, validated mitoRNAi effect with QPCR and northern blotting analysis, and ruled out mitochondrial pseudogene implication in nucleus via mitochondrial DNA-depleted cells (see Fig 1). Next we explained the strategy on knocking down complex subunits from perspective of stability gaps across mtDNA-encoded protein subunits, and further validated the knock-downs from perspective of changes in rate of nascent protein. Lastly, using mitoRNAi, we report significant impact on mitochondrial function by the mtDNA-encoded subunits.
To date, mitochondrial genome function has been studied in the mutation analysis, while mutant cells are subject to differential adaptation. Our design of mitoRNAi can rule out such adaptation, which can be extended to broader areas in mitochondrial genome function analysis. To illustrate the power of this tool, we used mitoRNAi to study the regulation mechanism on respiratory chains, which has provided a new tool and ideas for upcoming research. More importantly, mitoRNAi has the potential to offer potential treatment solutions on diseases induced by mtDNA mutation.
GAO Kuanxing, a doctoral student under the joint-doctoral program by Wuhan University and the CAS IBP, and CHENG Man, a doctoral student of CAS IBP, are co-first authors of the article. CAS researcher ZHANG Xiaorong and Prof. FU Xiangdong of the University of California, San Diego, are correspondence authors. The work was funded by grants from the Ministry of Science and Technology of China, the National Natural Science Foundation of China, and the CAS. The MitoOct reagents used in the ClickIn test was provided by Prof. Michael P. Murphy of Medical Research Council UK.
Article link: https://www.nature.com/articles/s41422-020-00394-5
(Reported by IBP's Mitochondrial Biology Team)

