SUN Fei's Group Publishes Paper on Molecular Mechanism of Mitochondrial Phosphatidate Transfer
Communications Biology published a joint research paper by SUN Fei team of the Institute of Biophysics (IBP), China Academy of Sciences (CAS) and FAN Jun team of City University of Hong Kong (CityU) entitled "Molecular mechanism of mitochondrial phosphatidate transfer by Ups1" on August 25, 2020. By studying the saccharomyces cerevisiae PA transfer complexes Ups1/Mdm35, they analyzed crystal structures of apo- and DHPA-bound Ups1/Mdm35. A model of Ups1/Mdm35-membrane interaction was established using combined crystallographic data, all-atom molecular dynamics simulations, extensive structural comparisons, and biophysical assays.
As a specific phospholipid in inner mitochondrial membrane (IMM), cardiolipin (CL) plays a key role in maintaining the form and functions of mitochondria. CL is synthesized at the matrix side of the IMM via an enzymatic cascade initiating with PA. And PA is predominantly synthesized in the endoplasmic reticulum and imported to the inner mitochondrial membrane (OMM) via endoplasmic reticulum-mitochondria encounter structure (ERMES). In 2010, German researcher Thomas Langer et al. reported mitochondrial intermembrane Ups1/Mdm35 and its mammalian homologous complexes PRELID1/TRIAP1 also play roles in PA transfer. After that, a succession of research papers on molecular mechanism of PA transfer was published. In spite of that, though researchers reported two states of Ups1/Mdm35–apo and substrate binding, it remains unknown the exact intermediary steps in PA transfer, such as how Ups1/Mdm35 is bound to the membrane, how PA enters binding pocket of Ups1, and how Ups1-PA complex dissociates from the membrane. This research analyzed crystal structures of Ups1/Mdm3 in three states, including domain-swapped, apo, and DHPA-bound (1,2-dioleoyl-sn-glycero-3-phosphate). Different from existing researches, the DHPA-bound state showed weak interactions between Mdm35 and Ups1. The team also conducted biophysical assays that showed for Ups1 α3-helix mutants, Ups1-membrane interactions were reduced, thus decreasing total membrane-bound Ups1 levels. Through all-atom molecular dynamics simulations and mutant assays, it was discovered that the α2-loop, L2-loop, and α3 helix of Ups1 mediate membrane interactions, and that W65 and F69 on α2-loopare alternatively inserted into the membrane during PA transfer to anchor Ups1 molecule. For the first time, the two teams proposed the molecular dynamics equation for Ups1/Mdm35-mediated PA transfer, and by derivation, the equilibrium constant function of Ups1/Mdm3–membrane binding. Lastly, based on theoretic derivation and assays, a model of Ups1/Mdm35-membrane interaction was established (see Fig 1), which helps enhance understanding on working mechanism of lipid transfer proteins.
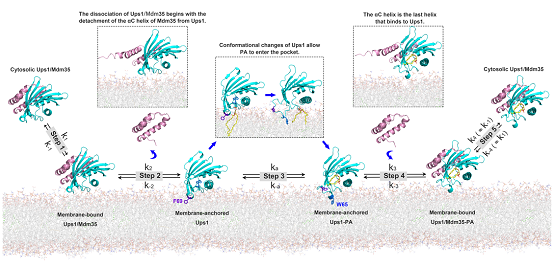
Fig 1: A working model for Ups1/Mdm35-mediated PA transport.
ZHAI Yujia (associate researcher) and SUN Fei (researcher) of CAS, Prof. FAN Jun of the CityU are co-correspondence authors. LU Jiuwei, YU Leiye (doctoral graduates of SUN's team) and Dr. Chun Chan of the CityU are co-first authors. Shanghai Synchrotron Radiation Facility collected diffraction data, and the Core Facility of Protein Sciences and Laboratory of Proteomics, Institute of Biophysics assisted in fluorescence spectrometry, mass spectrometry analysis, and NMR analysis. This work was supported by grants from Ministry of Science and Technology of China, Natural Science Foundation of China, and CAS.
See details in: https://www.nature.com/articles/s42003-020-01121-x
Contact: SUN Fei
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: feisun@ibp.ac.cn
(Reported by Dr. SUN Fei's group)

