Scientists Uncover the Structural basis for nucleosome-mediated inhibition of cGAS activity
On October 13, 2020, a research paper led by Dr. ZHANG Xinzheng and Dr. XU Rui-Ming at Institute of Biophysics (IBP), entitled "Structural basis for nucleosome-mediated inhibition of cGAS activity", was published online in Cell Research. In this work, the researchers report two cryo-EM structures of the complex of human cGAS (cyclis GMP-AMP synthase) and nucleosome, revealing the structural basis and regulatory mechanism of the inhibition of cGAS activity by the nucleosome, and help to understand the strategy by which hosts regulate cGAS activation to avoid autoimmune attack.
In the innate immune system, the cGAS-STING signal pathway plays a pivotal role in the defense of pathogen invasion or other cellular damages. cGAS is an important cytosolic sensor recognizing double-stranded DNA (dsDNA) generated by pathogen infection or cellular stress. Binding to dsDNA activates cGAS to synthesize the secondary messenger, cyclic GMP-AMP (cGAMP). Recognition of cGAMP by the adaptor protein stimulator of interferon genes (STING) further activates the downstream inflammatory responses, resulting in the production of type I interferons and other cytokines. However, cGAS is activated by dsDNA with no DNA sequence specificity, and aberrant activation of cGAS could lead to autoimmune diseases. Therefore, one interesting dilemma is how cGAS escapes from activation by self-DNA. Some studies suggest that cGAS is located only in the cytosol, but it is exposed to chromosomes during mitosis due to the breakdown of the nuclear envelop. Other studies pointed out that cGAS is located both in the nucleus and in the cytosol, and the nuclear cGAS is associated with chromatin. In both cases, the activity of cGAS is inhibited. Recently, a study showed that nucleosomes could competitively interact with cGAS and attenuate dsDNA-induced cGAS activation. However, the structural basis and detailed inhibitory mechanism for nucleosome-mediated attenuation of cGAS activity are still poorly understood.
Researchers from ZHANG Lab and XU Lab further demonstrated that nucleosome could competitively inhibit the activity of cGAS, and reported two cryo-EM structures of human cGAS bound to nucleosomes, with 1:1 and 2:2 binding stoichiometry, respectively (Figure 1). cGAS possesses three binding sites (site A, B and C) to mediate dsDNA interaction, and all of the three sites participate in the stimulation of cGAS activity. As shown in the structures of human cGAS-nucleosome complexes, cGAS bound to the nucleosome as a monomer, which interacts with the acidic patch of the H2A-H2B heterodimer through the region around site B, and contacts the DNA from the adjacent nucleosome via the basic residues within site C. Although site A dose not directly contact the nucleosome, it is not accessible to dsDNA due to steric hindrance. Therefore, the interaction between cGAS and the nucleosome prevents cGAS from further binding to dsDNA and locks cGAS to the inactive monomeric state, and thus blocking the dsDNA-induced stimulation of the cGAMP synthesis. Furthermore, researchers found that cGAS could interact with nucleosomes forming higher-order complexes, and successfully obtained a density map representing a 4:3 cGAS-nucleosome complex. Through a series of in vitro pull down assays, EMSA experiments and enzymatic activity assays, researchers examined the amino acids that participate in the inhibition of cGAS activity by the nucleosome, as well as the interaction regions important for the formation of the higher-order complex.
Prof. ZHANG Xinzheng and Prof. XU Rui-Ming are co-corresponding authors, Dr. CAO Duanfang, Dr. HAN Xiaonan and FAN Xiaoyi are co-first authors of this study. The work was funded by the National Key R&D Program of China, the National Natural Science Foundation of China, the Strategic Priority Research Program of the Chinese Academy of Sciences and the Youth Innovation Promotion Association of the Chinese Academy of Sciences.
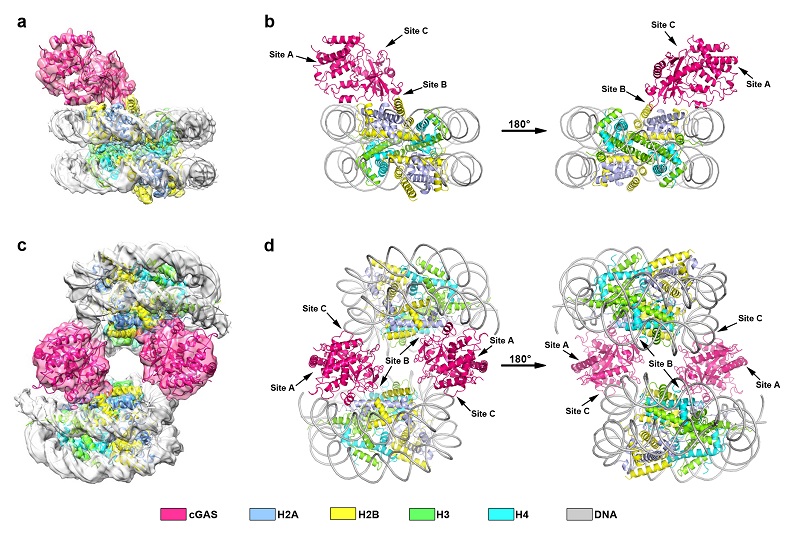
Figure 1. Overall structures of the cGAS-nucleosome complexes
(Image by Dr. ZHANG Xinzheng's group)
Article Link: https://www.nature.com/articles/s41422-020-00422-4
Contact: ZHANG Xinzheng
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: xzzhang@ibp.ac.cn
(Reported by Dr. ZHANG Xinzheng's group)

