Study Reveals Molecular Mechanism of Chromatin Loosening Induced by Open Nucleosomes
Researchers from the Institute of Biophysics of Chinese Academy of Sciences (CAS) published their findings entitled "Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2" in the journal THE EMBO JOURNAL on October 19, 2020.
Led by Prof. ZHOU Zheng and ZHU Ping, participants employed cryo-electron microscopy (Cryo-EM) to analyze nucleosome structures, which are composed of human histone variants such as H2A.B and H2A.Z.2.2, and explain nucleosome dynamics modulated by histone variant H2A as well as the molecular mechanism of chromatin structure.
A nucleosome consists of a core of histone proteins H2A, H2B, H3 and H4 and a double-stranded DNA with a length of 147bp, wrapping on the histone core in 1.6 laps. Nucleosomes are known for their stable structure and are insensitive to the formation of DNA and the transformation of histone-based decoration.
Histone variants can modify the transcriptional gene regulation of nuclesome and chromatin structures. The nucleosomes of histone H3 variants are the largest transformational CENP-A nucleosomes amongst all measured mono-nucleosomes. The length of DNA contained in CENP-A nucleosomes is 12 bp and there are no remarkable changes to the structure of protein cores.
H2A.B and H2A.Z.2.2, two variants of histone H2A, have displayed specific expression in spermatogonium and human brain. They play critical roles in spermatogenesis, transcriptional initiation and RNA splicing.
The open nucleosome structure, formed by H2A.B and H2A.Z.2.2, destroys the chromatin structure and leads to chromatin loosening. However, it is difficult for researchers to obtain the high-resolution structure of nucleosomes as they are in an unstable condition.
To get a stable condition of H2A.B, researchers rearranged and expressed peptide-binding domain of PARP1-DBD in a combination with DNA relevant with PARP1 and added it to H2A.B nucleosomes with the aim of producing Cryo-EM samples. Biochemical and EM findings show that PARP1-DBD can reduce the volatility of nucleosome DNA end but show no effect on nucleosome structure. They further interpreted the nucleosome structure of H2A.B histone vibrant with a resolution of 2.8 angstroms.
Compared with normal nucleosomes, the nucleosome of H2A.B variant has undergone noticeably transformational changes in its protein core and witnessed the shortage of some structural elements. The enlarged distance between H2A.B-H2B dimers has prolonged screw pitches, which wrap around DNA, and enhanced nucleosome thickness. Meanwhile, the length of DNA wrapped on histone cores has been shortened to 103 bp, reducing the laps to 1.2 when measuring how long DNA can bind protein cores.
Researchers also conducted an analysis of 3.9 angstroms EM of histone variant H2A.Z.2.2. They found that there are similarities between protein cores of H2A.Z.2.2 and H2A.B nucleosomes when it comes to their transformational changes and that the length of DNA surrounded protein core of H2A.Z.2.2 is roughly 125 bp. Both Mnase-sep and octamer packing experiments show that there is a regulating-octamer-folding (ROF) sequence of six consecutive amino acid residues in the C-domains of H2A.B and H2A.Z.2.2 and that the specific ROF sequence of H2A.Z.2.2 can greatly boost the efficiency of H2A.Z enzymatic transesterification reaction.
In sum, the research found the most open nucleosome structure. It also uncovered the structural changes of nucleosomes modulated by H2A.B and H2A.Z.2.2 and the molecular mechanism of chromatin dynamics.
The study was funded by the National Natural Science Foundation of China, the National Key Research and Development Program of China of the Ministry of Science and Technology, and the Strategic Priority Research Program of the CAS.
Prof. ZHOU Zheng and ZHU Ping are corresponding authors of the publication. ZHOU Min, an assistant researcher with Prof. ZHU, and DAI Linchang, a special research aid to Prof. ZHOU are first-authors of the research article. Other contributors include Dr. LI Chengmin, Dr. HUANG Yan and doctoral student SHI Liuxin.
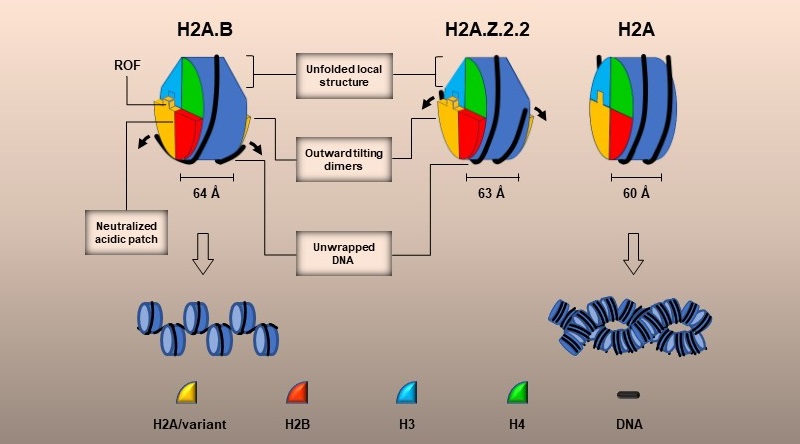
An illustration of the structure and structural changes of nucleosome variants
(Image by Dr. ZHOU Zheng's group)
Article Link: https://www.embopress.org/doi/full/10.15252/embj.2020105907
Contact: ZHOU Zheng
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: zhouzh@ibp.ac.cn
(Reported by Dr. ZHOU Zheng's group)

