Researchers Reveal Regulatory Mechanism of Retrotransposons in the Heterochromatin near the Centromeric Region
Heterochromatin refers to the DAPI-strained areas that are densely condensed in the genome. Previous studies suggested that heterochromatin genomes were the transcriptionally inactive state of chromatin. But with the development of transcriptomics sequencing technology, it has been discovered that heterochromatin is not always in a silent state.
It has been found in genomics studies that the stability of heterochromatin can ensure the stability of genome structure in cell interphase. The stability of heterochromatin, especially in and near the centromeric region, can facilitate the efficient and high fidelity transmission of genetic information during cell division. H3K9me3, a marker of constitutive heterochromatin, is randomly assigned to two daughter cells during cell division. The loss of H3K9me3 leads to loss of silencing of heterochromatin.
Repetitive sequences refer to the homologous DNA fragments that are present in multiple copies in the genome, which occupies a considerable proportion of the genome size, especially in heterochromatin. With multiple copies, it is difficult to find its accurate location in sequencing analysis, and the research on repetitive sequences has been lagging.
As the function of repetitive sequences has been reported in some known pathways, it has been found that repetitive sequences play their role accompanied by transcription. Therefore, the systematic study of the transcription of repetitive sequences and the binding sites of the RNA produced in these transcriptions will open a door for researchers to analyze the function of repetitive sequences in a systematic way.
FU Xiangdong, Professor of Cellular and Molecular Medicine at University of California, San Diego, in collaboration with Academician CHEN Runsheng and Prof. HE Shunmin of the Institute of Biophysics, Chinese Academy of Sciences (CAS), published a long cover story titled "Active retrotransposons help maintain pericentromeric heterochromatin required for faithful cell division" on Genome Research in November.
This research found that in Drosophila S2 cells, there are repeat RNAs that significantly bind to centromeric constitutive heterochromatin, which help maintain the state of heterochromatin and maintain the correct mitosis of cells.
By using GRID-seq technology (Global RNA interactions with DNA by deep sequencing) developed by the research group led by FU Xiangdong, Dr. HAO Yajing of the Institute of Biophysics, CAS (now a postdoctoral fellow in FU's research group at the University of California, San Diego) and other researchers improved the calculation methods, and systematically constructed a genome-wide map of repeat RNAs in S2 cells of Drosophila.
HAO and her teammates found for the first time that unlike the RNAs that tend to bind to active chromatin regions, a type of repeat-derived RNAs are significantly enriched in the constitutive heterochromatin regions. This type of RNA was subsequently named CHARRs (constitutive heterochromatin associated RNAs), which can both bind to neighboring sites in cis-acting and reach far through trans-acting. Interestingly, in regions where heterochromatin is activated, the more local RNAs are found, the less trans RNAs there are, and vice versa (Figure 1).
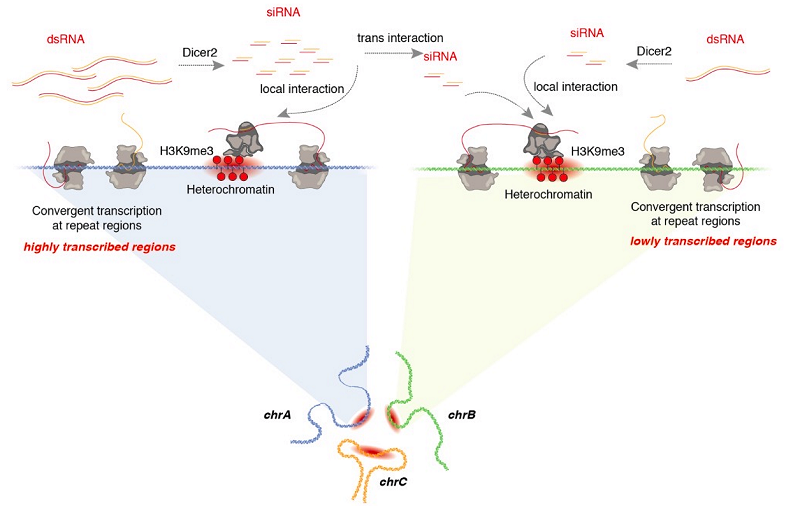
Figure 1. Stable mode of constitutive heterochromatin
(Image by Center for Big Data Research in Health)
The study also found that CHARRs are predominantly derived from active gypsy elements. In line with the characteristics of endo-siRNAs, they are about 21 nt in length and can be processed by Dcr-2 into small RNAs and bound by AGO2.
Previous reports including this article found that the knockdown of Dicer-2 can cause heterochromatin loss and abnormal chromosome arrangement and segregation during cell division. However, little is known about whether this key phenotype is caused by the siRNA pathway or other functions of Dicer-2 in the nucleus.
HAO reported for the first time that synthetic repeat-derived endo-siRNA mimics are sufficient to rescue Dcr-2-deficiency-induced defects in heterochromatin formation in interphase and chromosome segregation during mitosis. In addition, the recovery mechanism is sequence-specific.
The study demonstrates for the first time that endo-siRNA regulates the stability of constitutive heterochromatin in higher species, and it also demonstrates that active retrotransposons are required for stable genetic inheritance.
Meanwhile, the GRID-seq technology is proved to play a powerful role in the systematic study of RNA-chromatin interaction/RNA-chromatin maps as well as their functions.
FU Xiangdong, CHEN Runsheng and HE Shunmin are the co-corresponding authors of the paper; Dr. HAO Yajing of the Institute of Biophysics, CAS (now a postdoctoral fellow in FU Xiangdong’s research group at the University of California, San Diego) and Dr. WANG Dongpeng (now a postdoctoral fellow at the National Cancer Institute of the US National Institutes of Health) are the co-first authors of the paper.
ZHANG Peng, an associate professor and Dr. WU Shuheng of the Center for Big Data Research in Health of the Institute of Biophysics, CAS, participated in this research.
The project was funded by the National Natural Science Foundation of China, the National Key R&D Program of China, and the U.S. National Institutes of Health.
Article Link:https://genome.cshlp.org/content/30/11/1570.full
Contact: He Shunmin
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: heshunmin@ibp.ac.cn
(Reported by Center for Big Data Research in Health)

