YE Keqiong Laboratory made new progress in profiling RNA ribose methylation in Arabidopsis
On March 30, 2021, the YE Keqiong Laboratory at the Institute of Biophysics, Chinese Academy of Sciences (CAS), in collaboration with Dr WNAG Yuqiu at the School of Advanced Agricultural Sciences, Peking University and the LI Jiayang Laboratory at the Institute of Genetics and Developmental Biology, CAS, published a paper entitled "Profiling of RNA ribose methylation in Arabidopsis thaliana" in Nucleic Acids Research. This study systemically measured 2'-O-methylation modification in several RNAs in the model plant Arabidopsis thaliana and assigned guide snoRNA for modification sites.
Most RNAs undergo chemical modification after transcription. Methylation at the 2'-O position of ribose is one of the most frequent modifications. This type of modification occurs widely in various RNAs such as rRNA, tRNA and snRNA and modulates the structure, function and stability of RNA. In eukaryotes, cytoplasmic rRNA and snRNA possess abundant 2'-O-methylated nucleotides and majority of them are synthesized by box C/D small nucleolar RNA (snoRNA)-protein complexes. The C/D snoRNA in the complex recognizes substrates through base pairing interaction and selects a site located 5 nt upstream the box D/D' motif for modification (the D+5 rule).
In the model plant Arabidopsis thaliana, most C/D snoRNAs have been characterized but there is a lack of systemic mapping of 2'-O-methylation in RNA. The researchers profiled 2'-O-methylation in several Arabidopsis RNAs by the high-throughput RiboMeth-seq technique. They identified 111 modification sites in cytoplasmic rRNA, including 12 novel sites, and excluded many previously predicted false sites. They have assigned guide snoRNA for most sites using an updated snoRNA list and found that C/D snoRNAs frequently form extra pairs with nearby sequences, which may facilitate the substrate binding. By prediction and mutational validation experiments, they found that at least four modification sites are directed by guides with multiple specificities not using the D+5 rule.
They identified 19 modification sites in snRNAs, including 13 sites conserved in human. Chloroplast and mitochondrial rRNAs contain five almost identical methylation sites, including two novel sites mediating ribosomal subunit joining. They also analyzed the change of 2'-O-methylation in C/D snoRNP mutants. Overall, this study reveals the comprehensive 2'-O-methylation maps for Arabidopsis rRNAs and snRNAs and would facilitate study of their function and biosynthesis.
This work was resulted from collaboration between Institute of Biophysics, Chinese Academy of Sciences (CAS), the School of Advanced Agricultural Sciences, Peking University and the Institute of Genetics and Developmental Biology, CAS. Dr YE Keqiong is the corresponding author of the paper and graduate student WU Songlin, Dr WANG Yuqiu and Dr WANG Jiayin are the co-first authors. This work was supported by National Natural Science Foundation of China, Strategic Priority Research Program of Chinese Academy of Sciences and National Key R&D Program of China.
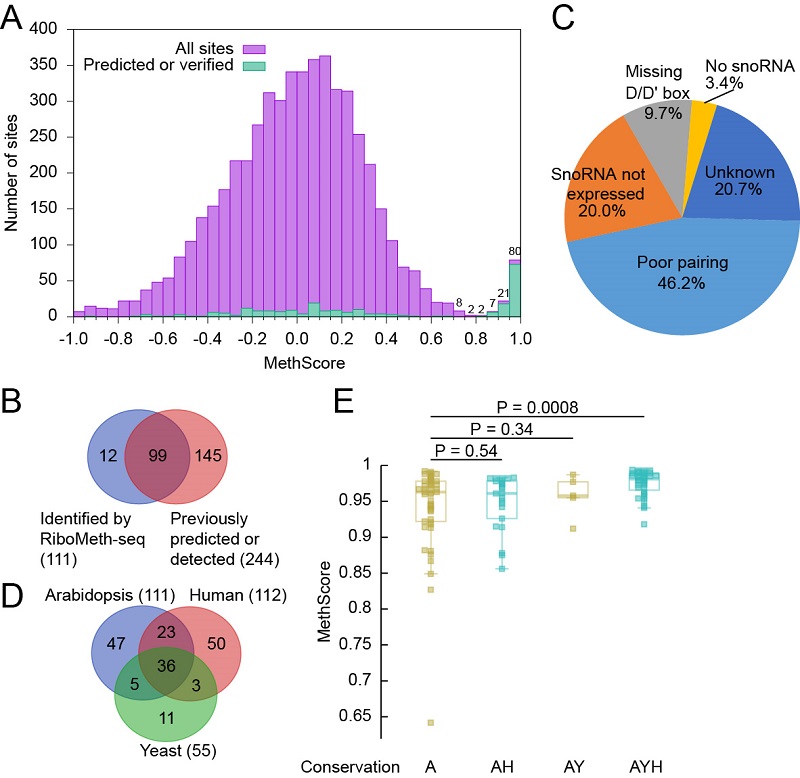
Figure. Profiling of 2'-O-methylation in Arabidopsis cytoplasmic rRNA.
Paper link: https://doi.org/10.1093/nar/gkab196
Contact: YE Keqiong
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: yekeqiong@ibp.ac.cn
(Reported by Dr. YE Keqiong's group)

