Scientists Develop New Interferometric 3D Single Molecule Localization Microscope
Precise localization of single molecules is essential for single molecule localization microscopy (SMLM) to investigate the 3D nanostructures in cells once concealed by the diffraction limit. However the axial resolution is mainly limited by the poor performance of axial localization methods based on point spread function (PSF) manipulations.
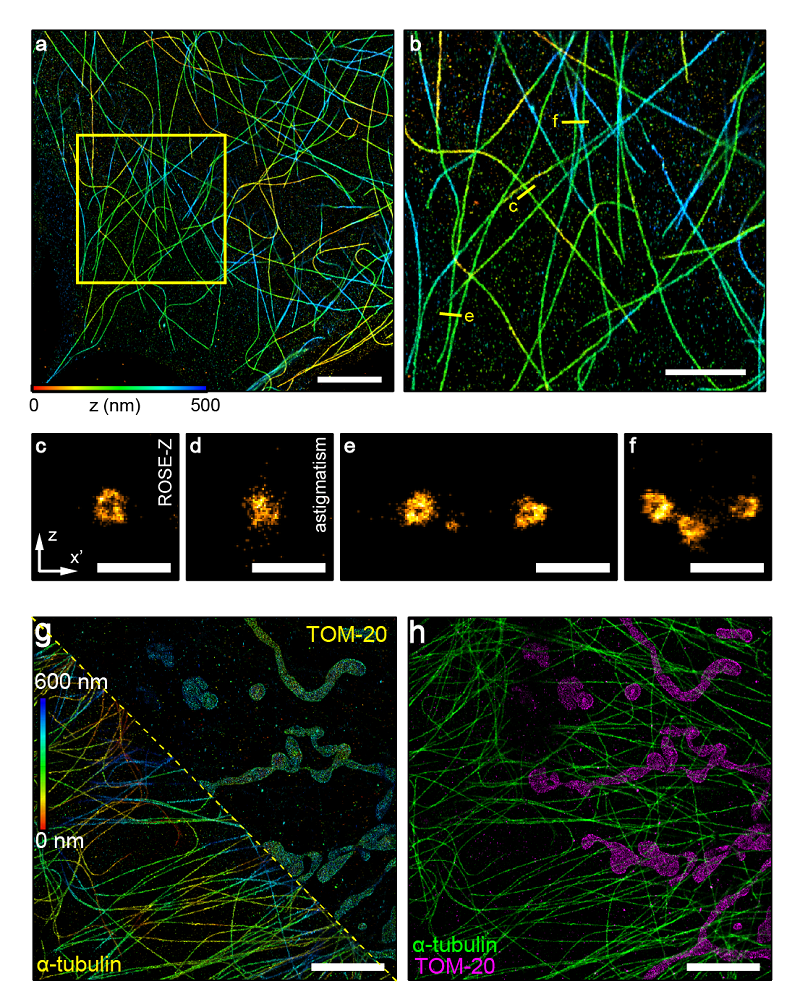
Recently, researchers from the Institute of Biophysics (IBP) of the Chinese Academy of Sciences (CAS) have developed a new axial localization with repetitive optical selective exposure (ROSE-Z) method for 3D super-resolution imaging. The work was published in Nature Methods online on April 1.
ROSE-Z method relys on an asymmetric optical scheme to achieve axial interferometric localization. Compared with the widely used astigmatism method, ROSE-Z yields approximately six-fold improvement on axial localization precision. The axial resolution of ROSE-Z was comparable or even better than lateral resolution, making ROSE-Z a feasible tool to study 3D nanostructures in cells.
Based on the advancement of ROSE-Z, <2 nm axial localization precision was achieved with only ~3,000 photons. The axial resolution is thus significantly improved, and researchers could be able to resolve the tubular structure of microtubule filaments in cells, which is hard to resolve by conventional methods.
Besides, two color imaging was demonstrated with ROSE-Z by adopting previously reported ratiometric methods with two far-red dyes: AF647 and CF660C. This method shows a low crosstalk below 1% between the two channels. This feature is convenient for studying the interaction between proteins or cellular structures.
Furthermore, thick sample imaging was tested and the result indicates uniformly high axial localization precision across the depth of field, up to 10 μm deep into the sample.
In short, this research provides a feasible and powerful tool to investigate the 3D nanostructures in cells, and shows its great potential in the biological research.
The article is available at https://dx.doi.org/10.1038/s41592-021-01099-2
Contact: JI Wei
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: jiwei@ibp.ac.cn
(Reported by Dr. JI Wei's group)

