Scientists uncover the structure basis for receptor recognition of Venezuelan equine encephalitis virus
On October 13, 2021, a research paper leaded by Dr. ZHANG Xinzheng at the Institute of Biophysics, Chinese Academy of Sciences and Dr. XIANG Ye at the School of Medicine, Tsinghua University, entitled "Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3" was published online in Nature. In this study, the researchers reported two cryo-EM structure of Venezuelan equine encephalitis virus (VEEV) and VEEV complexed with its receptor LDLRAD3 protein at 3.2 Å and 3.0 Å resolution, respectively, revealing the assembly mechanism of alphaviruses and VEEV host-cell receptor binding mechanism.
Venezuelan equine encephalitis virus (VEEV) is an RNA enveloped alphavirus that infects humans and equines, it can be transmitted by mosquitoes, jumping from animals to humans causing progressive central neurological disease and complications, sometimes lethal. Due to no vaccine or effective therapeutic drug for this virus, given its dangerous nature, VEEV is classified as a BSL3 virus. Recently, the receptor for VEEV was identified as the LDLRAD3 protein[1], however, the high-resolution three-dimensional structure of the VEEV virus and VEEV bound to its receptor haven't been revealed ,nor the specific molecular mechanism of the binding.
Researchers from Zhang Lab and Xu Lab further solved the structures of VEEV and VEEV complexed with LDLRAD3 at pseudo atomic resolutions using their previously developed "block-based" reconstruction method. The structures revealed that the extracellular region D1 of LDLRAD3 protein, inserted between the cleft formed by two adjacent spike-in proteins E1/E2 of VEEV by hydrophobic and polar interactions, binds at a molar ratio of 1:1. Interestingly, the same cleft that binds LDLRAD3 is also used by CHIKV, RRV, MAYV and ONNV for binding their host cell receptor Mxra8, both bind broadly similarly in the B domain of E2, but Mxra8 has a three amino acid bulge that prevents the formation of a salt bridge interaction with R64 of E2. Although the binding area is the same, critical binding sites of the two receptors are different, suggesting that VEEV enters the host via different binding receptor with other alphaviruses. As the first pseudo atomic resolution structure of VEEV, the structure also revealed the detailed interaction between the cytoplastic tail of glycoprotein E2 and the nucleocapsid protein. This interaction was believed to be essential for the nucleocapsid core to locate the glycoprotein shell of E1/E2 during alphavirus assembly and was never been resolved. Furthermore, mutagenesis study was performed on LDLRAD3 to verify the key residues involved in the interaction, in which the R41A mutation could enhance the binding ability of the receptor to the virus by approximately 10-fold, suggesting that LDLRAD3 could be used as a potential scaffold to study the inhibitor that blocks the cell host entry of VEEV.
This work benefited from the previously developed reconstruction method [2] in Prof. Zhang's lab. It is a continuation of the previous structural study of another Alphavirus [3]. Prof. Zhang and Prof. Xiang are the co-corresponding authors of the paper, and doctoral student MA Bingting in Xiang 's group, doctoral student HUANG Cuiqing and associate researcher MA Jun in Zhang 's group are the co-first authors of the paper. This work was supported by grants from the Ministry of Science and Technology of China and the Spring Wind Foundation of Tsinghua University, the National Natural Science Foundation of China, the Strategic Key Research Program of the Chinese Academy of Sciences, and the Frontier Science Key Research and Development Program of the Chinese Academy of Sciences, in addition to support from the Beijing Center for Frontier Research in Biological Structure, the Beijing Center for Advanced Innovation in Structural Biology, the Young Innovation Promotion Society of the Chinese Academy of Sciences, and the Beijing Science and Technology Rising Star Program.
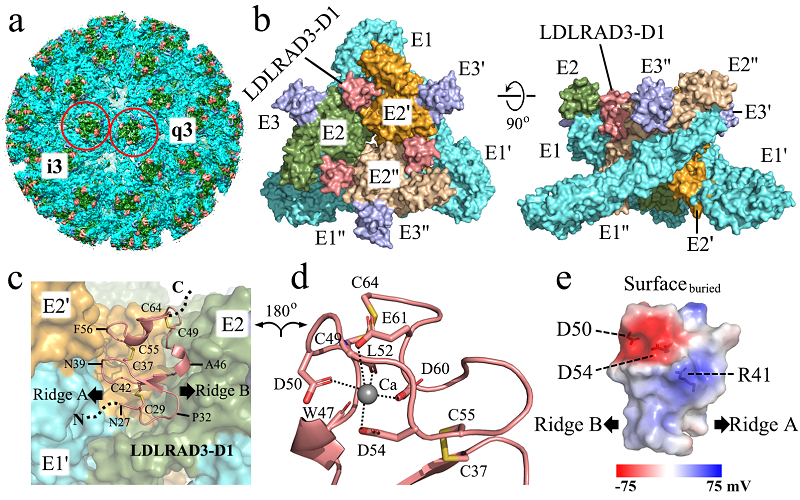
Figure1. Overall structure of the VEEV VLP-LDLRAD3-D1 complex and their detailed interactions.
Article link: https://www.nature.com/articles/s41586-021-03909-1
References
1. Hongming Ma, Arthur S Kim, Natasha M Kafai, James T Earnest, Aadit P Shah, James Brett Case, Katherine Basore, Theron C Gilliland, Chengqun Sun, Christopher A Nelson, Larissa B Thackray, William B Klimstra, Daved H Fremont, Michael S Diamond. LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature, 2020, 588(7837):308-314.
2. Dongjie Zhu, Xiangxi Wang, Qianglin Fang, James L Van Etten, Michael G Rossmann, Zihe Rao, Xinzheng Zhang. Pushing the resolution limit by correcting the Ewald sphere effect in single-particle Cryo-EM reconstructions. Nat Commun, 2018, 9(1):1552.
3. Lihong Chen, Ming Wang, Dongjie Zhu, Zhenzhao Sun, Jun Ma, Jinglin Wang, Lingfei Kong, Shida Wang, Zaisi Liu, Lili Wei, Yuwen He, Jingfei Wang, Xinzheng Zhang. Implication for alphavirus host-cell entry and assembly indicated by a 3.5 Å resolution cryo-EM structure. Nat Commun, 2018, 9(1):5326.
Contact: ZHANG Xinzheng
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: xzzhang@ibp.ac.cn
(Reported by Dr. ZHANG Xinzheng's group)

