Important progress in the study of the outer ring structure of Xenopus laevis (African clawed toad) nuclear pore complexes
On January 11, 2022, 《Protein & Cell》 published online a research article reporting the latest structural study of the nuclear pore complex (NPC) from Xenopus laevis entitled "8 Å structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI", achieved by Prof. SUN Fei's group of the State Key Laboratory of Biomacromolecules and Prof. ZHANG Chuanmao's group of Peking University. In this study, by combining the state-of-art techniques of cryo-electron microscopy (cryo-EM) and AI-based biomacromolecule structure prediction, researchers have obtained outer rings structure of X. laevis NPC at around 8 angstrom resolution, and a nearly complete pseudoatomic model of the outer rings was built upon. Other than well-established Y complex in the outer rings, in each asymmetric unit of the cytoplasmic ring, a pentameric Nup358 complex, two Nup214 complexes, two Nup205 and one Nup93 were distinguished. For nuclear ring, one ELYS, one Nup93 and one Nup205 were distinguished in each asymmetric unit. These results help to reveal the spatial position and interaction relationship between each nucleoporins in the outer ring of NPC, laying a foundation for further understanding of the function of the NPC.
Nucleus is the largest organelle in eukaryotic cells, and on the nuclear envelope there are pores with a biomacromolecule machine which mediate the cargo transportation and energy exchange between the nucleoplasm and cytosol, named as the NPC. From the cytoplasmic side to the nucleoplasm side, the architecture of NPC can be divided into cytoplasmic filament, cytoplasmic ring, inner ring, luminal ring, nuclear ring, and nuclear basket. The whole NPC presents a quasi-eight-fold symmetrical hollow cylindrical structure surrounding the central channel of the nuclear pore. In higher eukaryotes (such as humans, Xenopus, etc.), the molecular weight of NPC can reach more than 100 MDa. Resolving the high-resolution structure of the NPC is of great significance for understanding its assembly mechanism. However, due to its huge molecular mass and highly dynamic nature which adapt to its physiological functions, the NPC still lacks a reliable full atomic model, which limits the in-depth study of its structure and physiological function. In 2020, Prof. SHI Yigong's group from West Lake University used cryo-EM single particle analysis and tomography to study the structural characteristics of the cytoplasmic and luminal rings of the NPC from Xenopus laevis, which identified and located most secondary structure elements of cytoplasmic ring assemblies. In early January of 2022, Michael Rout's group in the United States published a paper in "Cell", which utilized comprehensive techniques to study the high-resolution structure of the yeast NPC in the purified state and the relatively low-resolution structure in its physiological state, elucidating the modularly assembled form of the yeast NPC and its relationship to physiological functional states. These studies have deepened the understanding of the mechanism of NPC assembly.
Prof. SUN Fei's group and Prof. ZHANG Chuanmao's group have further improved the cryo-EM single particle analysis technology according to the sample characteristics of X. laevis NPCs, to obtain various orientations for three-dimensional reconstruction. The final data processing results show that this strategy can achieve higher resolution than previous results using electron tomography, and most of the outer ring nucleoporins were resolved to secondary structure level. An overall resolution of around 8 angstrom can be achieved for different regions of the outer rings of the NPC.
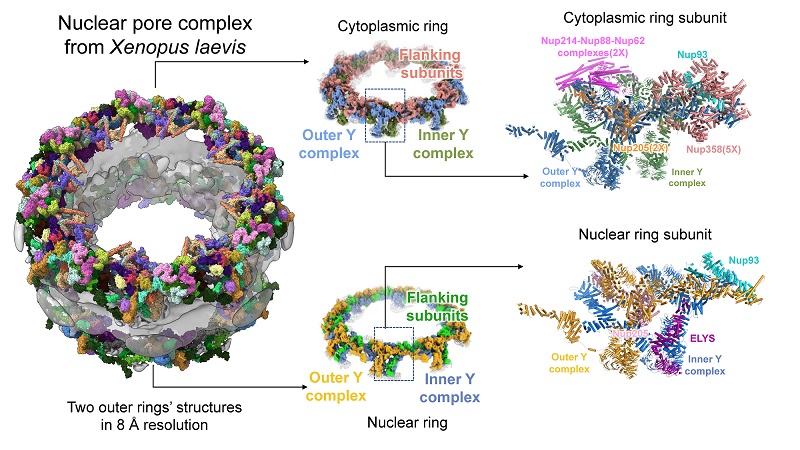
Figure 1. The fine structure of the outer ring of the Xenopus NPC reveals the composition and interaction patterns of numerous protein subunits.
On this basis, the researchers predicted the full-length three-dimensional structures of all nucleoporins in the Xenopus NPC with the help of the most accurate protein three-dimensional structure prediction software to date, named AlphaFold2. Then the most complete NPC outer ring structure model to date was obtained. This structure not only complements the missing part of the Y complex which is the essential backbone of outer rings (Fig. 1), but also identifies a series of nucleoporin subunits that play important functions on the outer ring. In each asymmetric unit of the cytoplasmic ring, a pentameric Nup358 complex was identified to stabilize the stem region of Y complexes; two Nup214 complexes were found to form the mRNP remodeling platform at the cytoplasmic side of the central channel; a Nup93 protein played a bridging role to connect the Y complexes; and two Nup205 proteins stabilize the head-to-tail manner of cytoplasmic ring structure. In each asymmetric unit of the nuclear ring, a Nup205 protein was identified to stabilize the nuclear ring structure; an ELYS protein initiates NPC assembly after mitosis; and a Nup93 bridges the stem region of the Y complex. These detailed structural information can provide important reference data for future studies on the assembly of NPCs.
Prof. SUN Fei from the Institute of Biophysics, Chinese Academy of Sciences, and Prof. ZHANG Chuanmao from Peking University, are the co-corresponding authors of this paper. Graduate student TAI Linhua and Prof. ZHU Yun from Prof. SUN Fei' group, and postdoctoral fellow Dr. REN He from Prof. ZHANG Chuanmao's lab are the co-first authors of this paper. This research was funded by the Strategic Priority Research Program of the Chinese Academy of Sciences, the National Natural Science Foundation of China and the grants from Ministry of Science and Technology of China. The sample screening of this study was completed in the Electron Microscopy Center, School of Life Sciences, Peking University, and the data collection was completed in the Center for Biological Imaging of the Institute of Biophysics. Special thanks give to Dr. HUANG Xiaojun of the Institute of Biophysics for her support and help in data collection. The research group of Prof. ZHANG Lihua, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, and the research group of Prof. FAN Jun, City University of Hong Kong also provided help for this research.
Contact: SUN Fei
Institute of Biophysics, Chinese Academy of Sciences
Beijing 100101, China
Email: feisun@ibp.ac.cn
(Reported by Dr. SUN Fei's group)

